
High mobility group box 2 modulates the progression of osteosarcoma and is related with poor prognosis
Introduction
Osteosarcoma (OS) is a common bone cancer that threatens the health and lives of children, teenagers, and young adults. Early hematogenous metastasis is detected in as many as 15–20% of patients at diagnosis (1). The lung is the most common site of distant metastasis, accounting for the major cause of osteosarcoma-related deaths (2). Newly diagnosed osteoasarcoma usually undertakes three to four cycles of neoadjuvant chemotherapy, which include cisplatin, doxorubicin and methotrexate (MTX), followed by surgical resection and additional chemotherapy after operation. Over the past three to four decades, these managements remain as major treatment toward OS patients. However, for patients with relapse or distant metastasis, the survival rate has seen little improvement since the 1980s. Substantial efforts have been made to gain a better understanding of OS and to help achieve better outcomes for OS patients.
As the second-most abundant class of proteins within cells, high mobility group (HMG) proteins play a global role in stabilizing the whole genome and act as important transcription factors (3). The HMG super-family, i.e., high mobility group box B (HMGB), includes 4 members HMGB1-4. HMGB1 and HMGB2 share identical amino sequence of up to 75%. Although the crucial role of HMGB1 in modulating tumorigenesis and progression has been reported in tremendous cancers, until recently the function of carcinogenesis and prognostic value of HMGB2 on malignant tumors was gradually uncovered (4). Patients with increased HMGB2 expression in glioblastoma, breast cancer, gastric cancer, and ovarian cancer have been reported to have lower 5-year and overall survival rates (5,6). By orchestrating the chromatin landscape of gene loci, HMGB2 can stimulate the transcription of metalloproteinases, anaerobic glycolysis-related kinase, and epithelial mesenchymal transition (EMT)-related proteins, thus promoting cancer cell proliferation and inducing distant metastasis (7,8).
Only a handful of studies have attempted to illustrate the role of HMGB2 in osteosarcoma. Analysis of gene expression microarray dataset GSE16089 including 3 samples of MTX-sensitive OS cell line SaoS2 and its counterpart MTX-resistant SaoS2 revealed HMGB2 as a potential regulator in osteosarcoma (9). However, the mechanism of HMGB2 modulation in OS is still unclear. In this study, we investigated the clinicopathological relationship between HMGB2 and OS. In vitro experiments were performed and the functions of HMGB2 were observed in OS cell lines. Further investigation was carried out using two mRNA microarray datasets, one miRNA microarray dataset, and one lncRNA dataset to explore the possible pathways in which HMGB2 might be involved in osteosarcoma and to explore the ceRNA network targeting HMGB2. By elucidating the biological action of HMGB2, we aimed to gain new insight into the regulation of HMGB2 in osteosarcoma.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4801).
Methods
Collection of clinical samples
A total of 78 osteosarcoma paraffin-embedded specimens were obtained from patients diagnosed as OS during 2010/05–2011/08 from the First Affiliated Hospital of Sun Yat-sen University (Guangzhou, China). Histopathological data and clinical information such as age, gender, tumor size, Enneking staging, distant metastasis, and survival were collected from these patients. In addition, 17 cases of benign bone tumor (osteoid osteoma) during the same period were collected. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Institutional Research Ethics Committee of our hospital approved the usage of the above-mentioned clinical materials for research purposes (No. [2017]232). Because of the retrospective nature of the research, the requirement for informed consent was waived.
Immunohistochemistry staining of HMGB2
Immunohistochemical (IHC) staining of the paraffin-embedded tissue sections was carried out to detect HMGB2 expression. Briefly, the sections were first deparaffinized and rehydrated, and then incubated in 3%H2O2 for 10 minutes. Then, antigen retrieval was conducted by heating the tissue sections in 10 mmol/L sodium citrate buffer (pH 6.0) for 15 minutes at 95 °C. Anti-HMGB2 (1:800, #ab11973, Abcam) was incubated at 4 °C overnight, phosphate-buffered saline (PBS) served as the negative control. Subsequently, the sections were incubated with secondary antibody (Dako, Carpinteria, CA, USA) for 30 minutes at room temperature. HMGB2 expression in the tissue sections was detected using non-biotin horseradish peroxidase detection system in line with the manufacturer’s protocol.
The expression of HMGB2 was assessed according to the intensity and percentage of positive cells. The intensity of HMGB2 expression was classified as negative [0], low [1], medium [2], or high [3]. The IHC score for HMGB2 expression was multiplied by the intensity and percentage of positive cells: IHC score = (intensity) × (percentage of positive cells). The cases were sorted by score into two groups: strong expression (a score of ≥180) and weak expression (a score of <180).
Cell cultures and treatment
The human osteosarcoma cell lines 143B and MG63 were obtained from the cell banks of our department and cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, Grand Island, NY, USA), with 10% FBS (Gibco, Grand Island, New York, USA), 100 µg/µL streptomycin, and 100 µg/µL penicillin in an incubator at 37 °C with 5% CO2.
The establishment of stable cell lines
To establish stable over-expression and knockdown cell lines in our study, 143B and MG63 cells were selected. Stable HMGB2 over-expression and knockdown cell lines were constructed (Genscript, Nanjing, China). The lentivirus PCD513B was purchased from Clontech. Briefly, HMGB2 over-expression as well as HMGB2 knockdown lentivirus vectors were constructed in 293T cells. HMGB2 coding sequence (NM_001130688.1) was amplified by polymerase chain reaction (PCR). Then, the product was cloned into vector and confirmed by sequencing. HMGB2 ShRNA (5'-CCG GAG TCC CTC TAA AGC AGC TCA ACT CGA GTT GAG CTG CTT TAG AGG GAC TTT TTT-3') was synthesized and cloned into plasmid (CDH-U6-ShRNA-EF1A-COPGFP-T2A-PURO) and confirmed by sequencing. Blank virus (PCDH-CMV-NEGATIVE-EF1A) was used as a control for both cell lines. The culture medium containing viruses was collected after 48–72 hours. Then, 143B and MG63 cells were transfected with the packed lentivirus and selected with 0.42 µg/mL puromycin 48 hours after transfection. The expression of HMGB2 was confirmed by real-time PCR and western blotting.
RNA extraction and quantitative real-time PCR
Total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. cDNA synthesis was carried out using 1.5–2 µg of RNA from each sample and the Revert Aid First Strand cDNA Synthesis kit (Roche, Diagnostic, Mannheim, Germany). Quantitative real-time PCR was conducted with FastStart Universal SYBR Green Master (ROX, Roche, Diagnostic, Mannheim, Germany). All experiments were performed in triplicate.
The primers used for amplification were:
HMGB2 (sense 5'-CGG GGC AAA ATG TCC TCG TA-3', antisense 5'-CGG AAG AGT CCG GGT GTT T -3');
GAPDH (sense 5'-GAC TCA TGA CCA CAG TCC ATG C-3', antisense 5'-AGA GGC AGG GAT GAT GTT CTG-3') (internal control).
Western blotting
Equal amounts of protein from each sample were loaded into SDS-polyacrylamide gel (10%) and then transferred onto a polyvinylidene fluoride (PVDF) membrane. The membrane was incubated with anti-HMGB2 (1:1,000, Cell Signal Technology, #14163), anti-GAPDH (1:3,000, Millipore Corp, #2118s, MA, USA) at 4 °C overnight, and then with secondary antibody accordingly. The protein bands were detected in enhanced chemiluminescence (ECL) and analyzed with a chemiluminescence imaging analyzer (ImageQuant LAS 4000, GE Healthcare, USA). The intensity of protein expression was quantified with Image J.
Cell proliferation analysis assay
Cell Counting Kit 8 (CCK8, Keygent, Suzhou, China) was used to analyze cell proliferation. Briefly, cultured cells were seeded in a 96-well plate at a cell count of 3×103/200 µL. At 12, 24, 48, and 72 hours, 10 µL of CCK8 solution was added to each well. After incubation for 4 hours, the absorbance was measured at a wavelength of 450 nm. Each experiment was performed in triplicate.
Cell migration and invasion assays
Cell migration and invasion assays were carried out using 24-well Biocoate cell culture inserts (Costar, Corning, NY, USA) equipped with polyethylene terephthalate membrane (8-µm porosity). For the cell invasion assay, wells were coated with Matrigel Basement Membrane Matrix (1:6, diluted with serum-free RMPI 1640 medium; Trevigen, Gaithersburg, MD, USA). The lower chamber of each well was filled with 10% FBS cell culture medium. The cells were re-suspended in serum-free DMEM and seeded into the wells at a total cell count of (0.5–2)×104. After incubation at 37 °C for 24–48 hours, the filters were fixed with methyl and stained with methyl violet. Cells remaining on the upper layer of the filter were removed, and those infiltrating the lower layer of the filter were counted under a microscope. All experiments were conducted in triplicate.
Would healing assay
Wound healing assays were conducted in a 6-well plate. Scratches were made in each well with a pipette tip. The cells were washed with PBS to remove the debris and then cultured in serum-free medium. Images of the wounds were captured under an inverted microscope at 0, 6, 12, 24, 36, and 48 hours to document cell migration. The wound area and healed wound percentage were analyzed with Image J.
Colony formation assay
The cells were trypsinized and then seeded in a 60-mm dish with a total cell count of 200 cells per dish. After 14 days of culture, the colonies were stained with 1% crystal violet. Afterward, the number of visible colonies were scored. All experiments were performed in triplicate.
Flow cytometry analysis
Flow cytometry was carried out to measure the apoptotic rate of OS cells. Apoptosis analysis was conducted using Annexin V-FITC kit (Keygent, Suzhou, China), following the manufacturer’s instructions. In brief, the cells were re-suspended in 500 µL Bing Buffer, then 5 µL Annexin V-FITC was added, followed by 5 µL propidium iodide (PI). After 5–15 minutes of incubation in darkness at room temperature, flow cytometry was conducted to measure the apoptotic rate of cells at a wavelength of 488 nm. All analyses were performed in triplicate.
Statistical analysis
The relationship between HMGB2 expression and clinical features was analyzed using chi-square tests (Pearson’s chi-square test and Yates’s correction for continuity) and Fisher’s exact test. Correlations between variables were calculated by Pearson’s correlation coefficients or Spearman’s rank correlation coefficients. Survival curves were estimated by the Kaplan-Meier method and the log-rank test was applied to compare the curves. Logistic regression and Cox proportional hazards regression were used for univariate and multivariate analysis. Data from the cell proliferation, migration, cell count, and apoptosis experiments as well as the relative protein expression of western blot were analyzed by two-tailed Student’s t-test or analysis of variance (ANOVA). All statistical analyses were performed using the Statistical Package for the Social Sciences version 25.0 for Windows (IBM Corp., Armonk, NY, USA) and R software for Windows version 3.6.1 (http://www.r-project.org/). A P value of <0.05 was considered statistically significant.
Bioinformatic analysis
Data were obtained from Gene Expression Omnibus (GEO) and our own microarray assay. Briefly, one miRNA microarray data set (GSE28423) and two mRNA microarray data sets (GSE11414 and GSE42352) were obtained from the GEO database of the National Center for Biotechnology Information (NCBI). GSE28423 (miRNA microarray data set) included 19 samples of osteosarcoma cell lines and 4 normal bone tissue samples. GSE11414 (mRNA microarray data set) included 4 samples of osteosarcoma cell lines and 2 normal osteoblastic cell line samples. GSE42352 (mRNA microarray data set) included 19 samples of osteosarcoma cell lines and 3 normal osteoblastic cell line samples. A LncRNA microarray, which included 6 samples of osteosarcoma cell lines and 3 normal osteoblastic cell line samples, was also conducted in our study.
All data were analyzed with principal component analysis and a boxplot on quality control and groupings. Then, the limma and affy packages were employed to analyze the differentially expressed RNA (DE_RNA). The pretreated data were set with a threshold of |log2(FC)|≥0.75, and a P value of <0.05 was considered to show significant differential expression. The DE_RNAs were presented and visualized with a heat map and a volcano map.
The MiRcode database was used to predict the target miRNAs of differentially expressed LncRNAs (DE_LncRNAs). These predicted target miRNAs were intersected with the differential miRNAs (DE_miRNAs) to enroll all possible DE_miRNAs. After that, miRanda, miRMap, and miRTarBase were employed to predict the target mRNAs of these DE_miRNAs, which were then intersected with differentially expressed mRNAs (DE_mRNAs) to enroll all possible DE_mRNAs. Base on the thesis that lncRNAs competitively bind with miRNAs to regulate mRNA expression with shared miRNA response elements, competing endogenous RNA (ceRNA) network was constructed and visualized with Cytoscape 3.4.0 (Figure 1). HMGB2 was specifically paid attention to during the conduction of analysis.
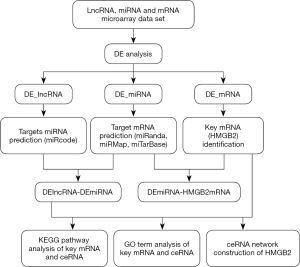
The clusterProfiler package and DAVID v6.8 were used to analyze the functions of DE_mRNAs in Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, especially those involving HMGB2 in osteosarcoma. A P value of ≤0.05 was considered to represent significant enrichment.
Results
Expression of HMGB2 and its clinicopathological features in osteosarcoma
HMGB2 immunohistochemical (IHC) staining was performed on 78 osteosarcoma and 17 osteoid osteoma tissue samples. HMGB2 was localized in the cytoplasm and nuclei of tumor cells. The overall expression of HMGB2 was stronger in the osteosarcoma tissues than in the osteoid osteoma tissues (Figure 2A). Strong HMGB2 expression was present in 40 of the 78 osteosarcoma samples (51.28%), while only 5 of the 17 osteoid osteoma samples (29.41%) had strong HMGB2 expression (P<0.05) (Figure 2B).
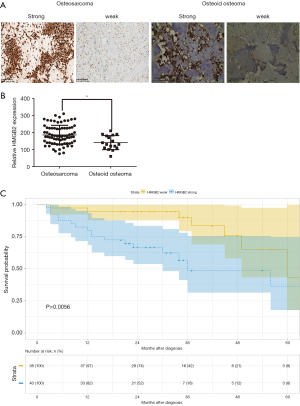
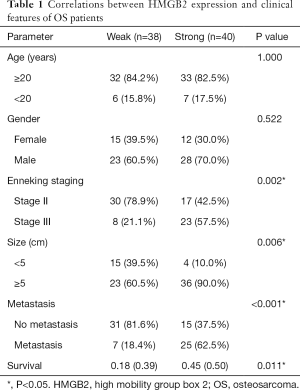
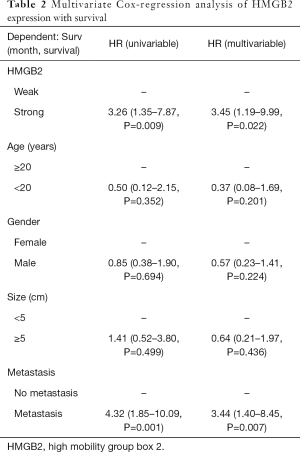
Statistical analysis was carried out to investigate the relationship between clinicopathological features and HMGB2 expression in OS patients. HMGB2 expression was found to be significantly associated with Enneking staging (P=0.002), tumor size (P=0.006), metastasis (P<0.001), and survival (P=0.011) (Table 1). Logistic regression analysis showed that strong HMGB2 expression might present as a risk factor for larger tumor size (OR =6.06, 95% CI: 1.90–23.68, P=0.004)), high Enneking stage (OR =5.28, 95% CI: 1.98–15.39, P=0.001), and metastasis (OR =7.96, 95% CI: 2.87–24.92, P<0.001). Multivariate Cox regression survival analysis demonstrated the independent prognostic value of strong expression of HMGB2 (HR =3.45, 95% CI: 1.19–9.99, P=0.022) after adjustment for age, gender, tumor size, and metastasis (Table 2). Kaplan-Meier survival curve analysis showed that the survival rate of OS patients with strong HMGB2 expression was significantly reduced compared with that of patients with weak HMGB2 expression. Further, the survival times of patients with strong and weak HMGB2 expression were significantly different (P=0.0056) (Figure 2C). Taken together, increased expression of HMGB2 in OS patients might be related to poor prognosis.
Full table
Full table
HMGB2 promoted proliferation and colony formation, and reduced apoptosis in osteosarcoma cell lines
To investigate the effect of HMGB2 on osteosarcoma cell lines, stable HMGB2 over-expression cell lines and stable HMGB2 knockdown cell lines were established and confirmed by western blot and RT-PCR (Figure 3A,B,C).
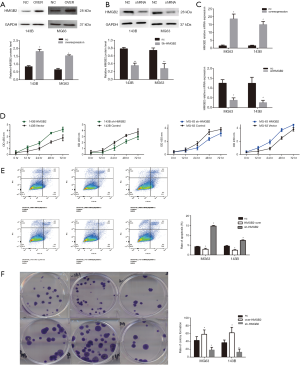
The CCK-8 assay revealed that, when compared with control group, over-expression of HMGB2 promoted cell proliferation (P=0.001), while knockdown of HMGB2 reduced the proliferation rate of OS cells (P<0.05). HMGB2 over-expression lowered the proportion of apoptotic cells 1.72–1.8 folds compared with the control group (P<0.05), whereas knockdown of HMGB2 increased the apoptotic rate 1.75–3.7 folds compared with the control group (P<0.05). Meanwhile, the colony formation assay revealed that HMGB2 over-expression increased the number of colonies formed 1.39–1.68 folds when compared with the control group, and HMGB2 knockdown led to a 2.36–2.9 folds decrease in colony formation when compared with the control group (P<0.05; Figure 3D,E,F).
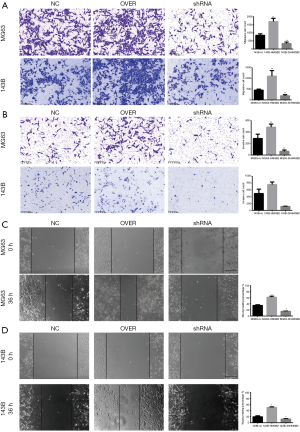
HMGB2 enhanced the migratory ability and invasive potential of OS cell lines
HMGB2 up-regulated the migratory ability and the invasive potential of OS cells. When compared with the control groups, cell migration was increased by approximately 2–2.3 folds in the HMGB2 over-expression cell lines (P<0.05), while knockdown of HMGB2 revealed a 2.3–5.1 folds decrease in cell migration (P<0.05) (Figure 4A). HMGB2 over-expression slightly increased the invasive capability of OS cells compared with the control group (P<0.05), whereas knockdown of HMGB2 markedly lowered the invasive capability of OS cells compared with the control group (P<0.05) (Figure 4B). The wound healing assay also indicated that HMGB2 over-expression improved the migration of OS cells when compared with the control group, while knockdown of HMGB2 inhibited the migration of OS cells (P<0.05) (Figure 4C,D).
HMGB2 modulates osteosarcoma via versatile pathways and might serve as a bona fide target gene of miR139-5p
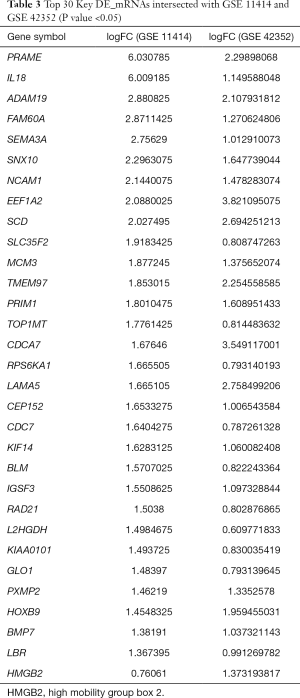
A total of 448 co-expressed DE_mRNAs were identified from GSE11414 and GSE42352, including HMGB2. Some of the key DE_mRNAs are listed in Table 3. A total of 268 DE_miRNAs (141 up-regulated miRNAs and 127 down-regulated miRNAs) were obtained from GSE28423. DE_lncRNAs in OS and osteoblastic cell lines were analyzed using the limma and affy packages, and eventually, 144 differentially expressed lncRNAs were obtained (89 up-regulated lncRNAs and 55 down-regulated lncRNAs).
Full table
The MiRcode database was used to predict the target miRNAs of DE_LncRNAs, which were then intersected with DE_miRNAs, and eventually, 1,149 miRNA-LncRNA pairs were obtained. The target mRNAs of the DE_miRNAs were predicted with miRanda, miRMap, and miRTarBase, and were then intersected with DE_mRNAs; eventually, 17 miRNA-mRNA pairs were obtained. Based on the thesis of ceRNA, a final ceRNA network including 63 DE_mRNAs was established.
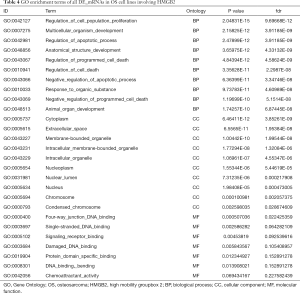
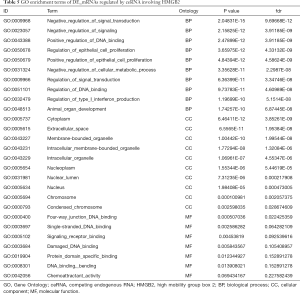
GO and KEGG analysis were performed to explore the pathways related to all DE_mRNAs and DE_mRNAs regulated by ceRNAs. Pathways involving HMGB2 among the top 10 most significantly enriched GO terms were listed in Tables 4 and 5. In biological process (BP) GO analysis, HMGB2 was found to be involved in 8 of the top 10 significantly enriched pathways of all DE-mRNAs and 5 of the top 10 significantly enriched pathways of DE_mRNAs regulated by ceRNAs. In cellular component (CC) GO analysis, HMGB2 was shown to be involved in 4 of the top 10 significantly enriched pathways of all DE-mRNAs and 2 of the top 10 significantly enriched pathways of DE_mRNAs regulated by ceRNAs (Figure 5A,B,C,D). BP terms revealed HMGB2 to be involved in the regulation of cell proliferation, apoptosis and programmed cell death, cell death, and anatomical structure development. CC items elicited possible functions of HMGB2 in the cytoplasm, nucleolus, membrane-bounded organelle, nucleus, and chromosome. Molecular function (MF) terms identified pathways such as DNA binding, protein receptor binding, protein domain-specific binding, and chemo-attractant activity.KEGG enrichment analysis did not show pathways involving HMGB2 (Figure 5E,F).
Full table
Full table
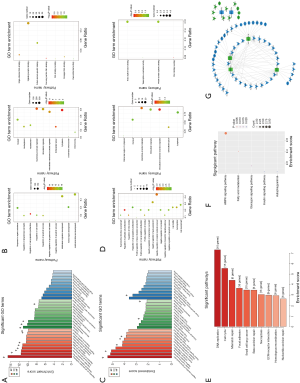
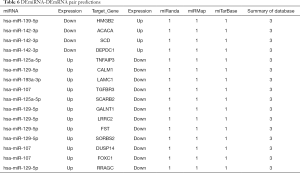
Next, the possible ceRNA regulation of osteosarcoma involving HMGB2 was explored. As mentioned above, 17 miRNA-mRNA pairs were obtained. HMGB2 was predicted as a bona fide target gene of miR139-5p, 6 candidate lncRNAs (MAGI1-IT1, DLX6-AS1, CPB2-AS1, PHF14, CCNYL2, and WHAMMP3) were elicited as having potential to competitively bind with miRNA139-5p and might form a ceRNA network with HMGB2 (Figure 5G, Table 6).
Full table
Discussion
OS is a highly malignant tumor which threatens the lives of patients despite adequate therapy. Since chemotherapy and surgery are the main treatments for osteosarcoma, patients with chemo-resistance and local relapse often encounter poor outcomes (10). Collaborative research with bioinformatic analysis has allowed researchers to gain a better understanding of the development of the disease and identify new biomarkers for prognosis, which will hopefully pave the way for new treatments and better outcomes for osteosarcoma patients (11,12).
Research has revealed the role of HMGB2 in maintaining normal function in the osseous system. HMGB2 expression in cartilage might facilitate joint movement and disperse mechanical pressure via the β-catenin pathway (13). A lack of HMGB2 may lead to osteoarthritis-like lesions (14). By regulating lymphoid enhancer-binding factor 1 (LEF1)-dependent transactivation, HMGB2 was shown to be responsible for chondrocyte enlargement through runt-related transcription factor 2 and the Wnt signaling pathway (15). Hence, in this study, we aimed to investigate the expression of HMGB2 in bone tumors and to explore its function in osteosarcoma cells.
In our study, we found that the expression of HMGB2 was higher in osteosarcoma tissues than in osteoid osteoma tissues. Strong expression of HMGB2 in osteosarcoma tissue was related to metastasis, tumor size, advanced stage, and patient survival. Multivariable Cox analysis revealed that increased HMGB2 expression might serve as a potential prognostic factor for osteosarcoma patients, which indicates that over-expression of HMGB2 might be related to poor outcomes of OS. Next, we tested the function of HMGB2 in OS cell lines. In vitro study demonstrated that over-expression of HMGB2 could promote cell proliferation and colony formation in OS cell lines, and enhance cellular migration and invasive capability, at the same time as reducing the apoptotic rate of cells. Meanwhile, knockdown of HMGB2 reduced cellular proliferation and colony formation, lowered the migration and invasive ability, and elevated the proportion of apoptotic cells. These results suggest that HMGB2 may play a crucial role in the tumorigenesis and progression of osteosarcoma. Previous studies have shown that in the nucleus, HMGB2 might act as a promoter complex with transcription factor TF IID-TF IIA (16). In cytoplasm, HMGB2 has been shown to act as a partner of assembly protein SET in nucleosome assembly (17). In our study, IHC examination revealed that HMGB2 was localized in both the cytoplasm and the nucleus, indicating that HMGB2 might play various roles in modulating osteosarcoma. Further in vivo and in vitro investigations are still needed to obtain a better understanding of HMGB2 in the modulation of osteosarcoma.
Based on the DE_mRNA enrichment results, we carefully explored the possible pathways in which HMGB2 might be involved. HMGB2 was shown to be involved in 8 of the top 10 significantly enriched BP GO terms and 4 of the top 10 significantly enriched CC GO terms. In the BP terms, HMGB2 was involved in the regulation of cell proliferation, apoptosis and programmed cell death, cell death, and anatomical structure development. These findings were consistent with those of previous studies that have demonstrated that HMGB2 can promote cell viability and invasion of cancer cell lines, indicating that HMGB2 could also be involved in critical processes of osteosarcoma progression (18). CC items analysis revealed that HMGB2 is located in the cytoplasm, nucleolus, membrane-bounded organelle, nucleus, and chromosome, and molecular function term analysis illustrated HMGB2’s involvement in DNA binding, protein receptor binding, protein domain-specific binding, and chemo-attractant activity. HMGB2 was reported to have multiple functions in the previous studies, including as a transcription-complex component, through collaborating with other proteins in organelle assembly, or through activating membrane receptor such as receptors for advanced glycation end products to induce artery injury (19). Our results elicited the possibility that HMGB2 also has versatile functions in regulating osteosarcoma through various pathways. However, we failed to identify KEGG pathways involving HMGB2, which is probably due to a lack of comprehensive analysis and verification of HMGB2. Therefore, it would be of high research value to gain a more in-depth perspective of HMGB2 on osteosarcoma regulation in future efforts.
Next, we attempted to identify whether HMGB2 is regulated by miRNAs or lncRNAs in osteosarcoma. Previously, a few miRNAs have been reported as targeting HMGB2. In Kaposi’s sarcoma, the lnc-OIP5-AS1/miR-218-5p network was shown to up-regulate HMGB2 expression and promote the proliferation and cell motility of HUVEC cells (20). The LncMALAT1/miR-1297/HMGB2 axis might play a crucial role in the process of tumorigenesis and progression of gastric cancer cell lines MKN45 and MKN28 (21). In melanoma, miR-329 was shown to arrest cell cycles, increase apoptosis, and impede cell proliferation and invasion, which are regulated by HMGB2 via the β-catenin pathway (22). In glioma cells, HMGB2 was proved to be downstream target of miR-130a, with the latter negatively regulating cellular proliferation (23). In IgA nephropathy (IgAN), miR-590-3p down-regulated HMGB2 expression in peripheral mononuclear cells and correlated with the severity of IgAN (24).
In our study, miR139-5p and six candidate lncRNAs were elicited with the expectation that they might form a lncRNA-miRNA network in HMGB2 modulation. MiR139-5p was reported to have a tumor suppression function when miRNA profiles in osteosarcoma, endometrial adenocarcinoma, breast cancer, and bladder cancer were explored (25-28). Down-regulation of miR139-5p was demonstrated to be related to persistent or recurrent thyroid cancer and responsible for tumor progression. An in vitro study showed that miR139-5p restrained cell proliferation and migration by regulating the abundance of the alternative splicing factor HNRNPF, which modulated genes involved in major cancer-related pathways (29). Among the six candidate lncRNAs, two lncRNAs were reported to be deregulated in different malignant tumors. LncRNA DLX6-AS1 promoted proliferation and metastasis of osteosarcoma through the miR641/HOXA pathway, or interacted with miR129-5p and increased the stemness of osteosarcoma via Wnt/β-catenin signaling activation (30,31). LncRNA MAGI1-IT1 is a newly identified lncRNA that was reported to promote proliferation and invasion in non-small cell carcinoma of the lung and epithelial ovarian carcinoma via binding with different miRNAs (32,33). However, whether these candidate lncRNAs might interact with miR139-5p and target HMGB2 in osteosarcoma still calls for in-depth investigation and verification.
There are some limitations to our study. First, the clinical sample size needs to be expanded so as to obtain more representative statistical data and to stratify patients with detailed clinical features, such as HMGB2 expression with sensitivity to chemotherapeutic treatment and its relevance with histological classification of osteosarcoma. Secondly, only limited analysis was carried out regarding the possible mechanism and pathway of HMGB2 in modulating osteosarcoma. Further in vitro and in vivo investigations should be undertaken to achieve a better understanding of HMGB2 and its down-stream pathway, as well as its ceRNA regulation network, in future.
Conclusions
In conclusion, we investigated the role of HMGB2 in osteosarcoma regulation. Clinically, increased HMGB2 expression might be related to poor outcomes among OS patients. In vitro experiments revealed that HMGB2 over-expression promoted cell proliferation and enhanced the migration and invasion abilities of cells. Also, we revealed that HMGB2 is involved in more than half of the top 10 pathways in GO term analysis, which indicates the versatile functions of this protein. Finally, in an attempt to explore the possible lncRNA-miRNA network targeting HMGB2, we identified miRNA139-5p as potentially targeting HMGB2. These results suggest the importance of HMGB2 in osteosarcoma progression and emphasize the necessity for future study.
Acknowledgments
Funding: This research was supported by the National Natural Science Foundation of China (Grant No. 81702655), Science and Technology Planning Project of Guangdong Province, China (Grant No. 2017A020215155), and the Natural Science Foundation of Guangdong, China (Grant No. 2018A030313551).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-4801
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-4801
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4801). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Institutional Research Ethics Committee of our hospital approved the usage of the above-mentioned clinical materials for research purposes (No. [2017]232). Because of the retrospective nature of the research, the requirement for informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Isakoff MS, Bielack SS, Meltzer P, et al. Osteosarcoma: Current Treatment and a Collaborative Pathway to Success. J Clin Oncol 2015;33:3029-35. [Crossref] [PubMed]
- Gao E, Li Y, Zhao W, Zhao T, et al. Necessity of thoracotomy in pulmonary metastasis of osteosarcoma. J Thorac Dis 2019;11:3578-83. [Crossref] [PubMed]
- Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev 2005;15:496-506. [Crossref] [PubMed]
- Mukherjee A, Vasquez KM. Targeting Chromosomal Architectural HMGB Proteins Could Be the Next Frontier in Cancer Therapy. Cancer Res 2020;80:2075-82. [Crossref] [PubMed]
- Cui G, Cai F, Ding Z, et al. HMGB2 promotes the malignancy of human gastric cancer and indicates poor survival outcome. Hum Pathol 2019;84:133-41. [Crossref] [PubMed]
- An Y, Zhang Z, Shang Y, et al. miR-23b-3p regulates the chemoresistance of gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis 2015;6:e1766. [Crossref] [PubMed]
- Aird KM, Iwasaki O, Kossenkov AV, et al. HMGB2 orchestrates the chromatin landscape of senescence-associated secretory phenotype gene loci. J Cell Biol 2016;215:325-34. [Crossref] [PubMed]
- Han Q, Xu L, Lin W, et al. Long noncoding RNA CRCMSL suppresses tumor invasive and metastasis in colorectal carcinoma through nucleocytoplasmic shuttling of HMGB2. Oncogene 2019;38:3019-32. [Crossref] [PubMed]
- Yang XR, Xiong Y, Duan H, et al. Identification of genes associated with methotrexate resistance in methotrexate-resistant osteosarcoma cell lines. J Orthop Surg Res 2015;10:136. [Crossref] [PubMed]
- Saraf AJ, Fenger J M, Roberts RD. Osteosarcoma: Accelerating Progress Makes for a Hopeful Future. Front Oncol 2018;8:4. [Crossref] [PubMed]
- Dai P, He Y, Luo G, et al. Screening candidate microRNA-mRNA network for predicting the response to chemoresistance in osteosarcoma by bioinformatics analysis. J Cell Biochem 2019;120:16798-810. [Crossref] [PubMed]
- Taniguchi N, Caramés B, Kawakami Y, et al. Chromatin protein HMGB2 regulates articular cartilage surface maintenance via beta-catenin pathway. Proc Natl Acad Sci U S A 2009;106:16817-22. [Crossref] [PubMed]
- Shane Anderson A, Loeser RF. Why is osteoarthritis an age-related disease? Best Pract Res Clin Rheumatol 2010;24:15-26. [Crossref] [PubMed]
- Taniguchi N, Kawakami Y, Maruyama I, et al. HMGB proteins and arthritis. Hum Cell 2018;31:1-9. [Crossref] [PubMed]
- Shykind BM, Kim J, Sharp PA. Activation of the TFIID-TFIIA complex with HMG-2. Genes Dev 1995;9:1354-65. [Crossref] [PubMed]
- Fan Z, Beresford PJ, Zhang D, et al. HMG2 interacts with the nucleosome assembly protein SET and is a target of the cytotoxic T-lymphocyte protease granzyme A. Mol Cell Biol 2002;22:2810-20. [Crossref] [PubMed]
- Wu ZB, Cai L, Lin SJ, et al. High-mobility group box 2 is associated with prognosis of glioblastoma by promoting cell viability, invasion, and chemotherapeutic resistance. Neuro Oncol 2013;15:1264-75. [Crossref] [PubMed]
- He YH, Wang XQ, Zhang J, et al. Association of Serum HMGB2 Levels With In-Stent Restenosis: HMGB2 Promotes Neointimal Hyperplasia in Mice With Femoral Artery Injury and Proliferation and Migration of VSMCs. Arterioscler Thromb Vasc Biol 2017;37:717-29. [Crossref] [PubMed]
- Li W, Wang Q, Feng Q, et al. Oncogenic KSHV-encoded interferon regulatory factor upregulates HMGB2 and CMPK1 expression to promote cell invasion by disrupting a complex lncRNA-OIP5-AS1/miR-218-5p network. PLoS Pathog 2019;15:e1007578. [Crossref] [PubMed]
- Li J, Gao J, Tian W, Li Y, et al. Long non-coding RNA MALAT1 drives gastric cancer progression by regulating HMGB2 modulating the miR-1297. Cancer Cell Int 2017;17:44. [Crossref] [PubMed]
- Mo Y, Fang RH, Wu J, et al. MicroRNA-329 upregulation impairs the HMGB2/β-catenin pathway and regulates cell biological behaviors in melanoma. J Cell Physiol 2019;234:23518-27. [Crossref] [PubMed]
- Tang C, Yang Z, Chen D, et al. Downregulation of miR-130a promotes cell growth and epithelial to mesenchymal transition by activating HMGB2 in glioma. Int J Biochem Cell Biol 2017;93:25-31. [Crossref] [PubMed]
- Zhai Y, Qi Y, Long X, et al. Elevated hsa-miR-590-3p expression down-regulates HMGB2 expression and contributes to the severity of IgA nephropathy. J Cell Mol Med 2019;23:7299-309. [Crossref] [PubMed]
- Li H, Zhang K, Liu LH, et al. MicroRNA screening identifies circulating microRNAs as potential biomarkers for osteosarcoma. Oncol Lett 2015;10:1662-8. [Crossref] [PubMed]
- Hiroki E, Akahira JI, Suzuki F, et al. Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci 2010;101:241-9. [Crossref] [PubMed]
- Krishnan K, Steptoe AL, Martin HC, et al. miR-139-5p is a regulator of metastatic pathways in breast cancer. RNA 2013;19:1767-80. [Crossref] [PubMed]
- Ratert N, Meyer HA, Jung M, et al. miRNA profiling identifies candidate mirnas for bladder cancer diagnosis and clinical outcome. J Mol Diagn 2013;15:695-705. [Crossref] [PubMed]
- Montero-Conde C, Graña-Castro O, Martín-Serrano G, et al. Hsa-miR-139-5p is a prognostic thyroid cancer marker involved in HNRNPF-mediated alternative splicing. Int J Cancer 2020;146:521-30. [Crossref] [PubMed]
- Li RY, Chen LC, Zhang HY, et al. MiR-139 inhibits Mcl-1 expression and potentiates TMZ-induced apoptosis in glioma. CNS Neurosci Ther 2013;19:477-83. [Crossref] [PubMed]
- Zhang N, Meng X, Mei L, et al. LncRNA DLX6-AS1 promotes tumor proliferation and metastasis in osteosarcoma through modulating miR-641/HOXA9 signaling pathway. J Cell Biochem 2019;10:1002. [PubMed]
- Zhang RM, Tang T, Yu HM, et al. LncRNA DLX6-AS1/miR-129-5p/DLK1 axis aggravates stemness of osteosarcoma through Wnt signaling. Biochem Biophys Res Commun 2018;507:260-6. [Crossref] [PubMed]
- Zhang G, Chen HX, Yang SN, et al. MAGI1-IT1 stimulates proliferation in non-small cell lung cancer by upregulating AKT1 as a ceRNA. Eur Rev Med Pharmacol Sci 2020;24:691-8. [PubMed]
- Gao H, Li X, Zhan G, et al. Long noncoding RNA MAGI1-IT1 promoted invasion and metastasis of epithelial ovarian cancer via the miR-200a/ZEB axis. Cell Cycle 2019;18:1393-406. [Crossref] [PubMed]


