Radiomics: an overview in lung cancer management—a narrative review
Introduction
With the rapid development of genomics, proteomics, metabolomics, and powerful methods for analyzing large sets of data, precision medicine, with targeted, individualized treatment, has become an active field, especially for cancer therapy (1). As early as 2003, the relationship between tumor gene variations and response to radiation therapy was evident, and it was regarded as the prelude to radiogenomics (2). Changes at the molecular level are believed to be essential factors for clinical decisions. However, the spatial and temporal heterogeneity of tumors limit the repeated use of invasive biopsies for cancer patients (3,4). In contrast, medical imaging represents an ideal means to capture the tumoral heterogeneity via a non-invasive manner. The underlying gene-expression patterns are represented as specific imaging traits. Therefore, imaging features could serve as potential molecular surrogates to optimize the treatment selection and management of cancers (5-7)—a fact which has become a cornerstone of radiogenomics. In short, the major objective of radiogenomics is to decode the tumor-inherited phenotypes using noninvasive imaging (5,8,9). The initial concept of radiomics was put forward by Baumann M, a German oncologist, in 2003, which was only focused on the specific relationship between gene expressions and radiosensitivity of tumors and was named as “radio-genomics” (5).
The innovations in medical imaging over the past decades have allowed for the high-throughput extraction of image information by automatic software, and faciliated crucial progress in radiogenomics. Large-scale radiographic features held great promise in translating the genomic and proteomics patterns into macroscopic imaging data. This was supported by the fact that different radiographic findings, such as the distinctive contrast-enhanced and non-enhanced regions on computed tomography (CT) scans, could reflect the variable protein expressions of glioblastoma (10). Based on further advanced quantitative analysis, it was further found that tumor diagnosis, treatment response, and even survival could be predicted by imaging (11,12). Medical imaging’s great expansion from an ancillary diagnostic tool to a big data bank with a central role in precision medicine is synonymous with the rise of radiomics (13,14). In 2012, Lambin P, a Dutch researcher, introduced the scientific term of “radiomics” for the first time, and defined it as to extract a large number of image features with a high-throughput approach. This was updated in 2014 and was emphasized the automated (or semiautomated) extraction of great amounts of quantifiable information in radiographic images, as well as proposed the purpose of radiomics was to correlate the radiography and intrinsic heterogeneity, genetic characteristics or other phenotypes to improve the management of diseases (14).
Lung cancer is the most common cause of cancer-related deaths worldwide (15). The management of lung cancer entails a multitude of difficulties, from diagnosis, to treatment options, to prognosis evaluation (16,17). With the evolution of radiomics, images have become informative data, rather than simply visible pictures, and their integration into lung cancer treatment may represent a paradigm shift in the field. Here, we review radiomics and its clinical application in lung cancer management, and summarize the limitations and future prospects. Literature published or online ahead of print in Pubmed, Embase, Ebsco, Web of knowledge, Ovid and China Biology Medicine disc in English or Chinese from 2010 with any study design were collected. We hope our review of radiomics in lung cancer would provide an idea about the value of radiomics in clinical practice, and help professionals understand radiomics in a more informed way.
The highlights and innovation of our review includes the following three points. First, we followed the real clinical process of lung cancer diagnosis and treatment to summarize the application of radiomics, which are pulmonary nodule detection and classification, histopathology and genetics diagnosis of lung cancer, clinical staging of lung cancer, therapy response and prognosis of lung cancer. This idea is consistent with the clinical practice. Therefore, it is apt to for readers to understand the clinical values of radiomics in the whole management process of lung cancer. Second, we have read and summarized the latest literature to keep readers abreast of advances. Third, research results of our group have been added and our real experience would make this review more readable to some extent. We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4589).
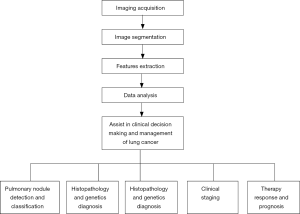
Workflow of radiomics
All modalities of digital imaging, including CT, magnetic resonance imaging (MRI), and positron emission tomography (PET), can be applied for radiomics analysis, with the workflow of radiomics across different types of imaging being essentially the same. But in the field of respiratory diseases, CT is the most commonly used modality. The main component of normal lung field is air with low density. When new lesions such as pulmonary nodules appear, local density increases and is significantly different from the surrounding normal tissues. The excellent density resolution of CT makes its wide use in pulmonary diseases, especially in pulmonary nodules.
Imaging acquisition
The first step of radiomics is to acquire high-quality, standardized imaging. However, the criteria for image acquisition in radiomics have not yet been established. It is recognized that CT scanning technical parameters (e.g., radiation dose, scanning protocol, with or without contrast agent, and so on) directly influence the radiomics features (18,19). Berenguer et al. confirmed that changes of imaging parameters could lead to the radiomics factors being nonreproducible (20). At present, images for radiomics analysis are mostly in Digital Imaging and Communications in Medicine (DICOM) format, which is obtained by reconstructing from the raw data of CT scanning. The convolution kernel and slice thickness were also found to affect the performance of a radiomics model to predict the epidermal growth factor receptor (EGFR) status in non-small cell lung cancer (NSCLC) (21). Moreover, post-processing can vary across vendors, and may thus yield different radiomic features (22). Therefore, it is hard to interpret results from studies based on the mixed heterogeneous cohorts with different CT parameters. Given these challenges, radiomic features must be reported with the image acquisition and reconstruction parameters.
Image segmentation
Image segmentation is a process that defines the region of interest (ROI) from the background or neighboring structures. It is usually achieved by the manual inspection of experienced investigators or the semiautomated/automated inspection by software. The greatest challenge for manual segmentation is interobserver variability and its time-consuming nature (23-26). Segmentation fully or partially guided by software is magnetic due to the improved efficiency, accuracy, reproducibility, and consistency this allows. Under certain circumstances, semiautomated/automated segmentation produces results highly similar to those of the manual approach in tumor volume measurement (27). However, when the tumor outline is unclear, manual segmentation is highly necessary.
Pulmonary nodule segmentation on CT images is based on detecting differences in density. The definite contrast between solid pulmonary nodules and perinodal normal lung tissues makes manual or semiautomated/automated segmentation simple. However, for subsolid pulmonary nodules, which includes pure ground glass nodules (pGGNs) and mixed ground glass nodules (mGGNs), segmentation is difficult, especially for the semiautomated/automated software, because of the vague margins (28). Difficulties also arise when drawing the outline of tumors affixed to the chest wall or mediastinum (29), and this might require hybrid methods of human–software interaction. Lacking this, most clinicians in pulmonary nodule studies continue to rely on accurate and reproducible manual segmentation (30-32). However, with the advent and development of deep learning, segmentation may be avoidable altogether, and an algorithm could be created to automatically extract features from an unsegmented image. This is one of the notable distinctions between radiomics and deep learning (33,34).
Features extraction
Quantitative imaging features of the ROI are extracted using high-throughput methods, and both two-dimensional (2D) and three-dimensional (3D) features can be derived. Data from 2D refers to the information on a single-slice image, while 3D indicates the entire volume of a tumor with many slices (22). In general, 3D radiomics features are more informative, and they are applied when the intratumoral heterogeneity needs to be explored. To a greater extent, 2D radiomics is used in a much easier and faster way. However, one study compared 2D and 3D CT radiomics features in predicting the prognosis of NSCLC and revealed that both 2D and 3D CT radiomic features have a certain prognostic ability. The authors of the study even recommended prioritizing 2D features (35). Therefore, for any given study, there is no definite option for the features selection.
The extracted radiomic features are usually categorized as semantic and agnostic (36). Semantic features are those features that are expressible via the radiologist’s lexicon, and include size, shape, intensity patterns, and so on; whereas features derived by computer such as spatial complexity and other texture information are agnostic features. Semantic features were reported to be powerful in predicting gene expression patterns in hepatocellular carcinoma and efforts are being made to capture such semantic data with the aid of computers to achieve higher prediction value (36). However, with the development of radiomics, more and more agnostic features, such as textural analysis, were evaluated, which were found to be useful in identifying certain clinical responses in lung adenocarcinomas but not all-powerful (37). Moreover, in our exploration for EGFR mutation prediction by radiomics, we found that all of the meaningful parameters were agnostic. However, Wang et al. compared the predictive performance of various methods in predicting the EGFR mutation in NSCLC, and results revealed that semantic model was not always significantly worse than other models (38).
Therefore, it might be regarded that both of semantic and agnostic features are indispensable in radiomics, and it is hard to say at this stage which is more important, even though agnostic ones seem to be more evident in some reports. At the same time, the algorithms, mathematical definitions, and nomenclature can affect the extraction results. Therefore, certain criteria for feature extraction and the standard group of features need to be first established. Lambin et al. recommended an image biomarker standardization initiative for quality control (39) and proposed a rigorous 16-component radiomics quality score (RQS) (40,41), which is technically required in radiomics research (42).
In theory, the number of radiomic features that can be extracted is almost unlimited, and depends on the filters, feature categories, and other parameters being used. If all possible features were included in a model, the result would be inevitably overfitted (40,41). Accordingly, extracted features are then moved into selection, which should be performed in a robust and transparent way. Features that have been identified as independent from other traits, reproducible, and prominent in the data should be selected, whereas variable features should be eliminated (42).
Data analysis
Selected features are then analyzed to establish models to provide risk stratification on clinical issues. There are several methods for data analysis, including support vector machine (SVM), logistic regression, random forest, Xgboost, gradient boosting decision tree (GBDT), and other methods (43,44). Modelling methods also have been shown to affect the prediction value (45), and each of them has distinct inherent limitations. For example, the disadvantage of logistic regression is independent assumption, and features appearing to be discretized in Bayesian networks (41). It is thus proposed that the performance of multiple modelling methodologies would ideally be compared in a specific study to choose the best one (41). Another principle for the choice of modelling method is that the work and results should be as reproducible as possible (41).
Application of radiomics in lung cancer
Pulmonary nodule detection and classification
The detection of small pulmonary nodules is time-consuming in daily practice and can oftern overlook these nodules, particularly if they are smaller than 5 mm in diameter or similar to the adjacent structures in density (46). In the National Lung Screening Trial (NLST), up to 8.9% of small lung cancers were not detected in the baseline scanning (47). There is great demand for computer-aided detection (CAD) tools to assist radiologists in nodule detection. In a proposed scheme, investigators used a nodular enhancement filter to segment the pulmonary nodule candidates and extract radiomic features, followed by a random forest method to quantify the heterogeneity and distinguish between the real nodules and false positive ones. The sensitivity of this scheme reached 88.9% with 4 false-positive detections per CT scan (48). More recently, promising results with increasing sensitivity for nodule detection have been reported (49,50).
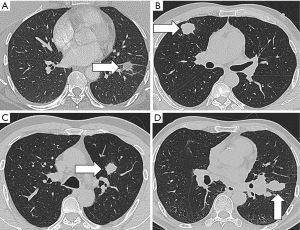
In addition to nodule detection, another important issue in CT scanning is the overdiagnosis of lung cancer. Results from the NLST indicated that only 3.6% of the detected nodules in the baseline screening were diagnosed as nascent lung cancer in the follow-up period, while the remaining 96.4% of nodules were overdiagnosed (51). This was based on the criteria in which nodules over 4 mm in size were defined as positive findings (52). Diagnostic approaches for defining lung nodules as benign or malignant usually rely on size and growth. Representative images for different pathological types of pulmonary nodules were showed in Figure 1. Radiomics provides a mass of information, making it possible to develop new techniques to address the accurate malignancy differentiation among detected nodules. Another study used 2 cohorts of screen-detected lung cancer and benign pulmonary nodules for image feature extraction in order to predict the subsequent emergence of lung cancer. A total of 219 3D radiomics features, describing the nodules’ location, size, shape, and texture were obtained. The 23 stable features were used in a random forests classifier to predict nodules that would become cancerous in 1 and 2 years after the screening. The accuracies in the training and validation cohorts were of 80% and 79%, respectively. These values were much better than those of the Lung Imaging Reporting and Data System and volume measurement, and were commensurate with the McWilliams risk assessment model.
A system that uses radiomics risk biomarkers to prescribe an optimal follow-up scheme for the detected lung nodules has been envisioned (53). Another deep learning methodology, in conjunction with an evolutionary technique and the particle swarm optimization algorithm, was developed to optimize the convolutional neural network for reducing the false-positive rate in lung nodule detection. For the Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IDRI), the accuracy of this model was 97.62%, while the sensitivity, specificity, and area under the receiver operating characteristic curve (AUC) were 92.20%, 98.64%, and 0.955, respectively (54).
The differential diagnosis of pulmonary part-solid nodules remains a clinical challenge. Digumarthy et al. carried out a retrospective study including 36 patients with 108 part-solid nodules, both with baseline and follow-up chest CT. The 3D radiomics features were extracted. Only 2 out of 92 radiomic features in the baseline scanning were able to differentiate the malignant from the benign nodules, with an AUC of 0.624; meanwhile, for the follow-up CT images, 52 out of 92 radiomic features could differentiate the 2 groups with an improved AUC of 0.708 (55). This study and others like it (56,57) demonstrate that radiomics has been embraced as an effective procedure for pulmonary diagnosis.
However, there are still a number of hurdles to overcome before these models can be widely applied in clinical practice. Subjects in the reported studies are from distinct cohorts, while there is variation in the original image data and in the methodologies implemented. Thus, the radiomic features and signatures are complicated and mutable. For example, in the study by Digumarthy et al., it was found that radiomics features in benign nodules were stable over time, whereas 63 out of 92 radiomics features of malignant nodules changed significantly between the baseline and follow-up chest CT scanning (55). Another study also indicated that the contrast-enhancement, reconstruction slice thickness and convolution kernel had a marked impact on the benign and malignant prediction of radiomics signatures (58). It is difficult to draw direct comparisons between the studies, and the value of this research is not yet directly applicable (59). Also, as the sample sizes of the datasets used have been relatively small, the scanner and scan protocols are insufficient, while the training and testing datasets are often not independent. The problem of overfitting is hard to avoid, and leads to difficulty in replicating the application of the models in separate datasets (39,60,61). Further research with much larger validated databases and comparable experimental protocols is likely needed.
Histopathology and genetics diagnosis of lung cancer
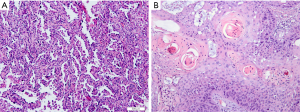
Histopathology subtypes, such as NSCLC (squamous cell carcinoma, adenocarcinoma) and non-small lung cancer (SCLC), are also critical for lung cancer treatment (Figure 2). One study extracted a total of 440 radiomic features from 2 independent lung cancer cohorts with a combined size of 350 patients. In the univariate analysis, 53 features were observed to be correlated with lung cancer histology, and the multivariate analysis revealed the highest performance of Naive Bayes classifier with 5 radiomic features (AUC: 0.72). This study highlighted the impressive potential of non-invasive and cost-effective radiomics in lung cancer histopathological classification (62). Similar results were achieved by Ferreira-Junior et al.’s quantitative radiomics analysis. In their study of 85 cases of lung cancer, they found that 45 features were associated with histopathology. A machine learning model yielded an AUC of 0.88 for prediction (63). Another multi-delineation analysis of 476 features in 4 cases of lung adenocarcinoma showed that robust features had high prognostic power in predicting the adenocarcinoma histology subtypes (64). Due to the remarkable heterogeneity of lung adenocarcinoma, there remains a need for the detailed classification of adenocarcinoma. For instance, in the classification criteria published by several oncology and respiratory societies, invasive lung adenocarcinomas were divided, primarily according to histologic features, into 5 pattern subtypes: lepidic, acinar, solid, papillary, and micropapillary (65). Micropapillary predominant lung cancers have been reported to have a poor prognosis (66). One study enrolled 339 patients with the determined amount of micropapillary component by the postoperative histologic examination. Radiomics analysis on the CT histogram, size, shape, gray-level co-occurrence matrix, intensity variance, and size zone variance revealed that intermediate grade, lower value of the minimum of the whole pixel, and lower value of the variance of the positive pixel, were predictors of the micropapillary component within lung adenocarcinoma. In the calibration plot, the combined radiomics features and clinical information yielded a prediction AUC of 0.751 (67).
Besides documenting the histopathology subtypes, genetic variation is also critical for clinical treatment modality. In a retrospective study, 219 3D CT radiomic features were extracted in 298 Asian patients with surgically resected peripheral lung adenocarcinoma, and 59 of these features were considered to be independent for further analysis. In addition to clinicopathological information (female, never smoking, lepidic predominant, and not highly differentiated), 11 radiomics features were found to be related with the EGFR mutation status. Finally, a multiple logistic regression model confirmed that a 5 radiomics signature, along with clinical data, to predict the EGFR mutation with an AUC of 0.709. This model thus represents a possible method for EGFR mutation prediction in lung cancer patients when profiling is unavailable (68). Our research group conducted a multicenter clinical trial in China to investigate EGFR mutation prediction as well. A total of 485 radiomic features reflecting the heterogeneity and phenotype of tumors from 180 lung adenocarcinoma patients were extracted. A least absolute shrinkage and selection operator (LASSO) based on multivariable logistic regression was used, and the radiomics features were found to have prognostic ability in predicting EGFR mutation status. The AUCs for the training and validation cohorts were 0.8618 and 0.8725 respectively, which were superior to the clinical variables alone (69). These results were repeated in the most recent studies with different machine learning methods (70,71). A group of 35 qualitative features extracted from PET/CT in 80 NSCLC patients with stage II and III before operation were also found to be significantly associated with EGFR mutation status. A predictive model based on the PET/CT data was established, and the prediction AUC was 0.953 (72). A radiomics signature might be useful in differentiating the EGFR-positive and wild types in lung adenocarcinoma.
Anaplastic lymphoma kinase (ALK) rearrangement is another driver mutation in NSCLC, and the echinoderm microtubule-associated protein-like 4 (EML4)-ALK fusion is regarded as the most common isoform (73). In a training set of 59 NSCLC patients with ALK positively rearranged, 24 CT traits plus 6 clinical-pathologic covariates were screened. Ultimately, 3 imaging traits, including central tumor location, absence of pleural tail, and large pleural effusion, with the age of patients younger than 60 years, were strongly associated with the positive ALK rearrangement. The sensitivity of the CT-radiomics-based model was 83.3% (15 of 18), and the specificity was 77.9% (74 of 95), with an accuracy of 78.8% (89 of 113) in the independent testing (74).
Crizotinib is recommended as the first-line treatment for positive ALK-fusion patients (75), and the success of recent research has elicited efforts to look for new oncogenic fusion genes, such as c-ros oncogene 1 (ROS1) and rearranged during transfection (RET) in NSCLC. A group of 539 pathologically confirmed lung adenocarcinoma patients with ALK/ROS1/RET fusion status were enrolled for radiomics analysis. Lower values for kurtosis and inverse variance on 3-voxel distance, combined with younger age, advanced tumor stage, and solid tumor on CT were applied to develop the prediction model for fusion status, yielding a sensitivity of 0.73 and a specificity of 0.70 (76). Other publications also produced similarly promising results in the genetics prediction of lung cancer (77-79). When biopsy tissues are unavailable, radiomics analysis may thus be a viable alternative.
Clinical staging of lung cancer
Clinical stage of tumors, which consists of the tumor (T), lymph node invasion (N) and distant metastasis (M), is decisive in selecting a treatment option. In the first report of radiomics in lung cancer, the correlation between radiomics and the TNM staging was identified (80). Our research group also explored the possibility of predicting distant metastasis in lung cancer based on CT radiomics features. Across 348 subjects, 485 radiomics features were extracted from pretherapy CT images. Concave minimization and SVM were used to select and evaluate the features’ ability. The AUC of this model, consisting of 4 radiomic features and 3 clinical factors, was 89.09%, which was far superior to that of the clinical-variable-only model (81). The results of our study were reproduced by Ferreira-Junior et al., who found that 40 and 2003 quantitative CT features were associated with lung cancer distant metastasis and nodal metastasis, with AUCs of 0.92 and 0.84, respectively (63). Beyond CT features, radiomics characteristics extracted from PET/CT, including statistical, histogram-related, morphologic and texture features, have also been able to quantify the intratumor heterogeneity of lung cancer. A prognostic model based on PET/CT radiomics features for predicting distant metastasis development was shown capable of distinctly separating the high- from the low-risk groups with more power than models based on the tumor volume or standard uptake value (SUV) (82). Another retrospective study on 545 cases even showed that PET texture features were capable of differentiating the primary lung cancer and metastatic lung lesions, but the model developed on CT features failed for predicting (83).
Standard lobectomy is the preferred treatment for most early stage lung cancer patients. Surgical dissection for lymph nodes should be avoided in subjects without nodal metastases (84). However, the accurate preoperative evaluation of lymph node metastases remains problematic. Although PET/CT is considered to be the best imaging modality, the false-negative rate and false-positive rate remain matters of concern (85,86), and new imaging methods must be developed. In one study, a group of 649 pre-surgical CT-based stage IA NSCLC patients were enrolled, 21% of whom had confirmed lymph node metastases after surgery, with 396 CT radiomic features and clinical information being collected. Receiver operating characteristic (ROC) analysis showed that the predictive value of the radiomics model for lymph node metastasis was 0.851. When combined with clinical data, the model’s prediction performance improved to 0.860. This study provides further evidence for the potential of presurgical CT-based radiomics in predicting lymph node metastases (87).
The brain is one of the most frequent sites of the extrathoracic metastasis of lung cancer. Once there is brain metastasis, the natural median progression-free survival (PFS) for patients reduces to 2 months (88). It is necessary to explore effective approaches in predicting the brain metastasis of lung cancer. In a study of 132 lung cancer patients with ALK-positive fusion in clinical stage III/IV, 1 radiomics feature of the pretreatment (but not during/after treatment) thoracic CT showed moderate discrimination ability for brain metastasis. This could be beneficial for risk stratification in advanced lung cancer patients (89). As for early-stage NSCLC, especially for category T1 lung adenocarcinoma in patients without any symptoms, there is still no consensus on the cost-effectiveness of routine screening for brain metastasis. If a robust prediction model for brain metastasis stratification with the visual assessment of thoracic CT were established, patients would benefit and the financial burden would be minimized. In another study, 89 eligible patients with category T1 lung adenocarcinoma were classified according to the presence of brain metastasis, and 1,160 radiomics features were extracted from the pretreatment unenhanced thoracic CT images. Prediction AUC of the radiomics model was 0.847, which increased to 0.871 when the model incorporated clinical information (90). Thus, thoracic CT-based radiomics analysis might be a promising approach for the clinical staging of lung cancer.
Therapy response and prognosis of lung cancer
Though the survival of lung cancer patients predominantly depends on the clinical stage, patients within the same stage exhibit wide variations in treatment response and outcome. Prognostic stratification is important for individualized management. An integrative analysis on 7 independent datasets with 1,194 NSCLC patients revealed that a 3D convolutional CT quantitative model could effectively predict the 2-year overall survival (OS) of patients treated by radiotherapy or surgery (91). In the recurrence-free survival (RFS) prediction for stage I NSCLC, 8 of 107 extracted CT features were used to develop a radiomics signature, which was helpful in stratifying patients into a high-risk (180/378) and low-risk group (198/378) with different RFSs (92). The response of SCLC patients to first-line chemotherapy was also investigated. A dataset including 92 patients receiving the standard first-line regimen of etoposide and cisplatin were divided into a response groups and no-response group. Twenty-one radiological features were used to establish a radiomics signature, and the AUC of its prediction performance was 0.797, which was better than that obtained by clinicopathological parameters. Therefore, CT-based radiomics has promise in chemotherapy response prediction as a guide for appropriate treatment planning in SCLC (93). Another study of 358 lung cancer patients extracted 665 3D radiomic features from PET/CT images using each of the 7 gray levels. The most predictive feature vector discovered (FVX) was independent of the other known factors and was invariant to the type of PET/CT manufacturer. Using the median cutoff, FVX showed power in predicting a 14-month survival difference in the validation cohort (94).
However, negative results have also been observed. In a longitudinal study including 141 NSCLC patients and 3 external datasets of 94, 61, and 41 for validation, no model for locoregional recurrence could be trained or validated (95). Other research on survival prediction of lung cancer patients using PET/CT features found that the AUC of the PET/CT-based radiomics model in external validation was no higher than 0.55 for any clinical endpoint, and robust independent PET/CT radiomics features could not predict the survival endpoints (96). Taken together, the findings above indicate that clinical decision and risk stratification based on either CT or PET/CT radiomics features is still problematic for lung cancer. Dataset heterogeneity and small cohort sizes might be the cause of poor prediction performance, and thus further studies are needed.
Precision medicine is particularly important for NSCLC patients in stage III, as local therapies (radiation or surgery) are not beneficial for all patients, while the value of adding chemoradiation to surgery remains unclear (97,98). A cohort of 85 patients with resectable local-advanced (stage II–III) NSCLC were included in a study. Radiomics analysis was performed on 85 primary tumors and 178 lymph nodes to predict pathological response after neoadjuvant chemoradiation before surgery. Three radiomic features, which described the primary tumor sphericity and lymph node homogeneity, were significantly predictive of pathological complete response with equal performance. Two features that quantify lymph node homogeneity could effectively predict the gross residual disease, and they performed much better than features of the primary lesions. This study suggests that lymph node phenotype presents an underlying sensitivity to chemoradiation and may provide a possible way to make an adaptive therapy plan for lung cancer patients (99).
Targeted therapy has been widely applied for lung cancer with driver gene mutations. It is well known that the patients with sensitive EGFR mutations can benefit from EGFR tyrosine kinase inhibitors (TKIs), including gefitinib, erlotinib, and afatinib (100,101). According to the National Comprehensive Cancer Network (NCCN), EGFR-TKIs are recommended as the first-line therapy for sensitive EGFR-mutated patients (102). However, PFS was found to vary across these patients from shorter than 1 month to several years, due to primary or secondary drug resistance (103,104). Even though biopsy is informative, individual assessment of the potential progression to EGFR-TKIs therapy remains challenging. In some cases, especially those in which the patient is too weak or the location of the tumor too unsuitable for biopsy, noninvasive diagnostic images provide a new approach for the risk stratification of drug response. In 2018, our group published a multicenter retrospective data study of what was then the first CT radiomics model to predict PFS survival in stage IV EGFR-mutant NSCLC patients with EGFR-TKI therapy. In total, 117 patients treated with EGFR-TKI were used as the training cohort, and 1,032 radiomic features were extracted from the pretreatment CT images. Twelve key features were screened out to develop the prediction model, and the performance was identified by the other three cohorts across three separate institutions. The AUC of this model was nearly 0.72 for the 10-month PFS prediction and 0.80 for the 1-year PFS prediction. The proposed prognostic strategy may be achievable via CT radiomics prediction, enabling us to conduct precise pretherapy management for EGFR-mutated NSCLC patients (105). A similar study was conducted to explore a CT-based radiomics signature as a prognostic factor in ALK-positive NSCLC with crizotinib therapy. In this research, 481 quantitative 3D features were derived from 63 stage IV cases. A radiomics model containing 3 features showed an effective value for the PFS prediction, with an AUC of 0.824 (106).
Immunotherapy has substantially changed the therapeutic strategies in lung cancer. Unfortunately, only a small portion of patients respond to treatment (107). There is an urgent need to develop methods to identify patients who are most likely to benefit from immunotherapy. The expressions of programmed cell death protein (PD)-1 and anti-programmed cell death ligand 1 (PD-L1) were at first encouraging, but later found to yield mixed results (108). In one study, CT-based radiomic features were extracted from 135 patients with advanced solid malignant tumors for training, 30 of whom had lung cancer. First, the tumor infiltration of CD8 cells was assessed by combining contrast-enhanced CT images and RNA-sequencing genomic data. A radiomics signature including 8 variables demonstrated an AUC of 0.67 for CD8 cell infiltration in the The Cancer Genome Atlas dataset for validation.
Sun et al. reported that in patients treated with immunotherapy, a higher baseline radiomics score was observed in cases with an objective response at 3 months and 6 months, with a much better OS. The results showed that radiomics could allow for the evaluation of tumor immune infiltration and thus be a novel predictor for the efficacy of immunotherapy (109). Another 203 patients with advanced melanoma and NSCLC undergoing anti-PD1 therapy were also analyzed, and 1,055 contrast-enhanced CT images for primary and metastatic lesions in pretreatment were collected. Quantitative features were used to distinguish between responding and nonresponding cases. The biomarker reached a significant performance on NSCLC lesions (AUC: up to 0.83), resulting in a 1-year survival difference of 24% (110). Furthermore, changes (“delta”) in the CT radiomic texture (DelRADx) before and after 2–3 cycles of immunotherapy were acquired from 139 NSCLC patients. A cluster of 8 DelRADx features was used to predict the immunotherapy response, yielding AUCs of 0.85 and 0.81 in 2 validation cohorts. Delta-radiomics risk-score might be another indicator for immunotherapy response (111).
Therefore, the workflow and clinical application of radiomics in lung cancer management were summarized in Figure 3.
Limitations and future prospects
Despite adding important information for clinical decisions, several limitations of radiomics should be also be addressed. The first major limitation is the lack of imaging criteria, which is the initial step of the radiomics workflow. As mentioned above, differences in scanning parameters, including dose administration, reconstruction kernels, and slice thicknesses, produce acquisition variability. Second, lesion segmentation has not been standardized to prevent the diversity in picking up features. For example, manual segmentation usually leads to variations in lesion boundary due to interobserver variability, while the process of automatic/semiautomatic segmentation is also not uniform. Third, the methods for feature selection are different across studies, making it exceedingly difficult for biomarkers to be reproduced. Finally, studies of radiomics have been retrospective in nature, and, although many correlations have been demonstrated, causality cannot be definitively confirmed.
These challenges notwithstanding, radiomics has enriched the research methods for exploring the biological behaviors of tumors. It is hoped the that the above-mentioned limitations will be gradually resolved in the future, with each of the workflow steps likely attaining standardization. Radiomics metrics, which are the least vulnerable to acquisition or reconstruction method flaws, might be explored in the future in more extensive applications. Moreover, the rapid progress of artificial intelligence will provide more tools for radiomics analysis, and facilitate its implementation, while the development of human-machine-assisted interpretation will make radiomics analysis more flexible and improve the accuracy of its results. Additionally, what are concerned is the role of radiologists and clinicians allied with radiomics. The work of radiologists and clinicians is not only to read and interpret images, but also to make comprehensive analysis and judgment in combination with other clinical information of patients. Many technical and legal problems in radiomics and artificial intelligence have not been solved yet. Therefore, it is unlikely that radiomics replace radiologists and clinicians in the near future. However, the development and application of radiomics will not only greatly improves the diagnosis level of primary medical institutions, but also greatly reduces the working pressure of doctors.
Conclusions
Here, we have reviewed the workflow and clinical utility of radiomics in lung cancer management, and remarked upon the potential future trends of this modality. A series of studies have shown the added value of applying radiomics analysis in the treatment of lung cancer. The implementation of radiomics is both feasible and invaluable, and has aided clinicians in ascertaining the nature of a disease with greater precision. However, it should be noted that radiomics in its current state cannot completely replace the work of therapists or tissue examination. More efforts are needed to overcome the limitations identified above in order to facilitate the widespread application of radiomics in the reasonably near future.
Acknowledgments
Funding: This study was supported by the National Key R&D Program of China Grant No. 2018YFC1311900 (to BC), the National Natural Science Foundation of China Grant No. 91859203 and No. 81871890 (to WL), and the Key Program from the Department of Science and Technology, Sichuan Province, China, Grant No. 2017SZ0052 (to BC).
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-4589
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4589). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Collins FS, Varmus H. A New Initiative on Precision Medicine. N Engl J Med 2015;372:793-5. [Crossref] [PubMed]
- Baumann M, Holscher T, Begg AC. Towards genetic prediction of radiation responses: ESTRO's GENEPI project. Radiother Oncol 2003;69:121-5. [PubMed]
- McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017;168:613-28. [PubMed]
- Hausser J, Alon U. Tumour heterogeneity and the evolutionary trade-offs of cancer. Nat Rev Cancer 2020;20:247-57. [Crossref] [PubMed]
- Rutman AM, Kuo MD. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol 2009;70:232-41. [Crossref] [PubMed]
- Harry VN, Semple SI, Parkin DE, et al. Use of new imaging techniques to predict tumour response to therapy. Lancet Oncol 2010;11:92-102. [Crossref] [PubMed]
- Gao X, Chu C, Li Y, et al. The method and efficacy of support vector machine classifiers based on texture features and multi-resolution histogram from (18)F-FDG PET-CT images for the evaluation of mediastinal lymph nodes in patients with lung cancer. Eur J Radiol 2015;84:312-7. [Crossref] [PubMed]
- Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol 2007;25:675-80. [Crossref] [PubMed]
- Gevaert O, Xu J, Hoang CD, et al. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology 2012;264:387-96. [Crossref] [PubMed]
- Hobbs SK, Shi G, Homer R, et al. Magnetic resonance image-guided proteomics of human glioblastoma multiforme. J Magn Reson Imaging 2003;18:530-6. [Crossref] [PubMed]
- Buettner R, Wolf J, Thomas RK. Lessons learned from lung cancer genomics: the emerging concept of individualized diagnostics and treatment. J Clin Oncol 2013;31:1858-65. [Crossref] [PubMed]
- Colen R, Foster I, Gatenby R, et al. NCI Workshop Report: Clinical and Computational Requirements for Correlating Imaging Phenotypes with Genomics Signatures. Transl Oncol 2014;7:556-69. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Chen B, Zhang R, Gan Y, et al. Development and clinical application of radiomics in lung cancer. Radiat Oncol 2017;12:154. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Cassim S, Chepulis L, Keenan R, et al. Patient and carer perceived barriers to early presentation and diagnosis of lung cancer: a systematic review. BMC Cancer 2019;19:25. [Crossref] [PubMed]
- Rai A, Doria-Rose VP, Silvestri GA, et al. Evaluating Lung Cancer Screening Uptake, Outcomes, and Costs in the United States: Challenges With Existing Data and Recommendations for Improvement. J Natl Cancer Inst 2019;111:342-9. [Crossref] [PubMed]
- Midya A, Chakraborty J, Gonen M, et al. Influence of CT acquisition and reconstruction parameters on radiomic feature reproducibility. J Med Imaging (Bellingham) 2018;5:011020. [Crossref] [PubMed]
- Kim H, Park CM, Lee M, et al. Impact of Reconstruction Algorithms on CT Radiomic Features of Pulmonary Tumors: Analysis of Intra- and Inter-Reader Variability and Inter-Reconstruction Algorithm Variability. PLoS One 2016;11:e0164924. [Crossref] [PubMed]
- Berenguer R, Pastor-Juan MDR, Canales-Vazquez J, et al. Radiomics of CT Features May Be Nonreproducible and Redundant: Influence of CT Acquisition Parameters. Radiology 2018;288:407-15. [Crossref] [PubMed]
- Li Y, Lu L, Xiao M, et al. CT Slice Thickness and Convolution Kernel Affect Performance of a Radiomic Model for Predicting EGFR Status in Non-Small Cell Lung Cancer: A Preliminary Study. Sci Rep 2018;8:17913. [Crossref] [PubMed]
- Hassani C, Varghese BA, Nieva J, et al. Radiomics in Pulmonary Lesion Imaging. AJR Am J Roentgenol 2019;212:497-504. [Crossref] [PubMed]
- Giraud P, Elles S, Helfre S, et al. Conformal radiotherapy for lung cancer: different delineation of the gross tumor volume (GTV) by radiologists and radiation oncologists. Radiother Oncol 2002;62:27-36. [PubMed]
- Van de Steene J, Linthout N, de Mey J, et al. Definition of gross tumor volume in lung cancer: inter-observer variability. Radiother Oncol 2002;62:37-49. [Crossref] [PubMed]
- Ito T. Effects of different segmentation methods on geometric morphometric data collection from primate skulls. Methods in Ecology and Evolution 2019;10:1972-84. [Crossref]
- Xu C, Howey J, Ohorodnyk P, et al. Segmentation and quantification of infarction without contrast agents via spatiotemporal generative adversarial learning. Med Image Anal 2020;59:101568. [Crossref] [PubMed]
- Rios Velazquez E, Aerts HJ, Gu Y, et al. A semiautomatic CT-based ensemble segmentation of lung tumors: comparison with oncologists' delineations and with the surgical specimen. Radiother Oncol 2012;105:167-73. [Crossref] [PubMed]
- Shakibapour E, Cunha A, Aresta G, et al. An unsupervised metaheuristic search approach for segmentation and volume measurement of pulmonary nodules in lung CT scans. Expert Systems with Applications 2019;119:415-28. [Crossref]
- Wang X, Cui H, Gong G, et al. Computational delineation and quantitative heterogeneity analysis of lung tumor on 18F-FDG PET for radiation dose-escalation. Sci Rep 2018;8:10649. [Crossref] [PubMed]
- Chen CH, Chang CK, Tu CY, et al. Radiomic features analysis in computed tomography images of lung nodule classification. PLoS One 2018;13:e0192002. [Crossref] [PubMed]
- Saba T, Sameh A, Khan F, et al. Lung Nodule Detection based on Ensemble of Hand Crafted and Deep Features. J Med Syst 2019;43:332. [Crossref] [PubMed]
- Tu SJ, Wang CW, Pan KT, et al. Localized thin-section CT with radiomics feature extraction and machine learning to classify early-detected pulmonary nodules from lung cancer screening. Phys Med Biol 2018;63:065005. [Crossref] [PubMed]
- Mariottoni EB, Jammal AA, Urata CN, et al. Quantification of Retinal Nerve Fibre Layer Thickness on Optical Coherence Tomography with a Deep Learning Segmentation-Free Approach. Sci Rep 2020;10:402. [Crossref] [PubMed]
- Zinchuk V, Grossenbacher-Zinchuk O. Machine Learning for Analysis of Microscopy Images: A Practical Guide. Curr Protoc Cell Biol 2020;86:e101. [Crossref] [PubMed]
- Shen C, Liu Z, Guan M, et al. 2D and 3D CT Radiomics Features Prognostic Performance Comparison in Non-Small Cell Lung Cancer. Transl Oncol 2017;10:886-94. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Phillips I, Ajaz M, Ezhil V, et al. Clinical applications of textural analysis in non-small cell lung cancer. Br J Radiol 2018;91:20170267. [Crossref] [PubMed]
- Wang S, Shi J, Ye Z, et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J 2019.53. [Crossref] [PubMed]
- Zwanenburg A, Vallieres M, Abdalah MA, et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020.191145. [Crossref] [PubMed]
- Park JE, Kim D, Kim HS, et al. Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement. Eur Radiol 2020;30:523-36. [Crossref] [PubMed]
- Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749-62. [Crossref] [PubMed]
- Dercle L, Lu L, Schwartz LH, et al. Radiomics Response Signature for Identification of Metastatic Colorectal Cancer Sensitive to Therapies Targeting EGFR Pathway. J Natl Cancer Inst 2020;112:902-12. [Crossref] [PubMed]
- Dev VA, Eden MR. Formation lithology classification using scalable gradient boosted decision trees. Computers & Chemical Engineering 2019;128:392-404. [Crossref]
- Zhou K, Jianyu Z, Ren Y, et al. A gradient boosting decision tree algorithm combining synthetic minority over-sampling technique for lithology identification. Geophysics 2020;85:1-52. [Crossref]
- Parmar C, Grossmann P, Bussink J, et al. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep 2015;5:13087. [Crossref] [PubMed]
- Rubin GD, Roos JE, Tall M, et al. Characterizing Search, Recognition, and Decision in the Detection of Lung Nodules on CT Scans: Elucidation with Eye Tracking. Radiology 2014;274:276-86. [Crossref] [PubMed]
- Scholten ET, Horeweg N, de Koning HJ, et al. Computed tomographic characteristics of interval and post screen carcinomas in lung cancer screening. Eur Radio 2015;25:81-8. [Crossref] [PubMed]
- Ma J, Zhou Z, Ren Y, et al. Computerized detection of lung nodules through radiomics. Med Phys 2017;44:4148-58. [Crossref] [PubMed]
- Yang Y, Feng X, Chi W, et al. Deep learning aided decision support for pulmonary nodules diagnosing: a review. J Thorac Dis 2018;10:S867-s75. [Crossref] [PubMed]
- Liu B, Chi W, Li X, et al. Evolving the pulmonary nodules diagnosis from classical approaches to deep learning-aided decision support: three decades' development course and future prospect. J Cancer Res Clin Oncol 2020;146:153-85. [Crossref] [PubMed]
- Manser R, Lethaby A, Irving LB, et al. Screening for lung cancer. Cochrane Database Syst Rev 2013;2013:CD001991. [PubMed]
- Black WC, Gareen IF, Soneji SS, et al. Cost-effectiveness of CT screening in the National Lung Screening Trial. N Engl J Med 2014;371:1793-802. [Crossref] [PubMed]
- Hawkins S, Wang H, Liu Y, et al. Predicting Malignant Nodules from Screening CT Scans. J Thorac Oncol 2016;11:2120-8. [Crossref] [PubMed]
- da Silva GLF, Valente TLA, Silva AC, et al. Convolutional neural network-based PSO for lung nodule false positive reduction on CT images. Comput Methods Programs Biomed 2018;162:109-18. [Crossref] [PubMed]
- Digumarthy SR, Padole AM, Rastogi S, et al. Predicting malignant potential of subsolid nodules: can radiomics preempt longitudinal follow up CT? Cancer Imaging 2019;19:36. [Crossref] [PubMed]
- Lee SH, Lee SM, Goo JM, et al. Usefulness of texture analysis in differentiating transient from persistent part-solid nodules(PSNs): a retrospective study. PloS One 2014;9:e85167. [Crossref] [PubMed]
- Wu W, Pierce LA, Zhang Y, et al. Comparison of prediction models with radiological semantic features and radiomics in lung cancer diagnosis of the pulmonary nodules: a case-control study. Eur Radiol 2019;29:6100-8. [Crossref] [PubMed]
- He L, Huang Y, Ma Z, et al. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep 2016;6:34921. [Crossref] [PubMed]
- Ather S, Kadir T, Gleeson F. Artificial intelligence and radiomics in pulmonary nodule management: current status and future applications. Clin Radiol 2020;75:13-9. [Crossref] [PubMed]
- Chalkidou A, O'Doherty MJ, Marsden PK. False Discovery Rates in PET and CT Studies with Texture Features: A Systematic Review. PLoS One 2015;10:e0124165. [Crossref] [PubMed]
- Choi W, Oh JH, Riyahi S, et al. Radiomics analysis of pulmonary nodules in low-dose CT for early detection of lung cancer. Med Phys 2018;45:1537-49. [Crossref] [PubMed]
- Wu W, Parmar C, Grossmann P, et al. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Front Oncol 2016;6:71. [Crossref] [PubMed]
- Ferreira-Junior JR, Koenigkam-Santos M, Magalhães Tenório AP, et al. CT-based radiomics for prediction of histologic subtype and metastatic disease in primary malignant lung neoplasms. Int J Comput Assist Radiol Surg 2020;15:163-72. [Crossref] [PubMed]
- Haga A, Takahashi W, Aoki S, et al. Classification of early stage non-small cell lung cancers on computed tomographic images into histological types using radiomic features: interobserver delineation variability analysis. Radiol Phys Technol 2018;11:27-35. [Crossref] [PubMed]
- Westaway DD, Toon CW, Farzin M, et al. The International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society grading system has limited prognostic significance in advanced resected pulmonary adenocarcinoma. Pathology 2013;45:553-8. [Crossref] [PubMed]
- Lee G, Lee HY, Jeong JY, et al. Clinical impact of minimal micropapillary pattern in invasive lung adenocarcinoma: prognostic significance and survival outcomes. Am J Surg Pathol 2015;39:660-6. [Crossref] [PubMed]
- Song SH, Park H, Lee G, et al. Imaging Phenotyping Using Radiomics to Predict Micropapillary Pattern within Lung Adenocarcinoma. J Thorac Oncol 2017;12:624-32. [Crossref] [PubMed]
- Liu Y, Kim J, Balagurunathan Y, et al. Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas. Clinical Lung Cancer 2016;17:441-8.e6. [Crossref] [PubMed]
- Zhang L, Chen B, Liu X, et al. Quantitative Biomarkers for Prediction of Epidermal Growth Factor Receptor Mutation in Non-Small Cell Lung Cancer. Transl Oncol 2018;11:94-101. [Crossref] [PubMed]
- Hong D, Xu K, Zhang L, et al. Radiomics Signature as a Predictive Factor for EGFR Mutations in Advanced Lung Adenocarcinoma. Front Oncol 2020;10:28. [Crossref] [PubMed]
- Jia TY, Xiong JF, Li XY, et al. Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modeling. Eur Radiol 2019;29:4742-50. [Crossref] [PubMed]
- Jiang M, Zhang Y, Xu J, et al. Assessing EGFR gene mutation status in non-small cell lung cancer with imaging features from PET/CT. Nucl Med Commun 2019;40:842-9. [Crossref] [PubMed]
- Zhao R, Zhang J, Han Y, et al. Clinicopathological Features of ALK Expression in 9889 Cases of Non-small-Cell Lung Cancer and Genomic Rearrangements Identified by Capture-Based Next-Generation Sequencing: A Chinese Retrospective Analysis. Mol Diagn Ther 2019;23:395-405. [Crossref] [PubMed]
- Yamamoto S, Korn RL, Oklu R, et al. ALK Molecular Phenotype in Non–Small Cell Lung Cancer: CT Radiogenomic Characterization. Radiology 2014;272:568-76. [Crossref] [PubMed]
- Deng H, Li B, Li L, et al. Clinical observation of crizotinib in the treatment of ALK-positive advanced non-small cell lung cancer. Pathol Res Pract 2019;215:152695. [Crossref] [PubMed]
- Yoon HJ, Sohn I, Cho JH, et al. Decoding Tumor Phenotypes for ALK, ROS1, and RET Fusions in Lung Adenocarcinoma Using a Radiomics Approach. Medicine (Baltimore) 2015;94:e1753. [Crossref] [PubMed]
- Wang T, Gong J, Duan HH, et al. Correlation between CT based radiomics features and gene expression data in non-small cell lung cancer. J Xray Sci Technol 2019;27:773-803. [Crossref] [PubMed]
- Lee G, Park H, Bak SH, et al. Radiomics in Lung Cancer from Basic to Advanced: Current Status and Future Directions. Korean J Radiol 2020;21:159-71. [Crossref] [PubMed]
- Tu W, Sun G, Fan L, et al. Radiomics signature: A potential and incremental predictor for EGFR mutation status in NSCLC patients, comparison with CT morphology. Lung Cancer 2019;132:28-35. [Crossref] [PubMed]
- Aerts HJWL, Velazquez ER, Leijenaar RTH, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Zhou H, Dong D, Chen B, et al. Diagnosis of Distant Metastasis of Lung Cancer: Based on Clinical and Radiomic Features. Transl Oncol 2018;11:31-6. [Crossref] [PubMed]
- Wu J, Aguilera T, Shultz D, et al. Early-Stage Non-Small Cell Lung Cancer: Quantitative Imaging Characteristics of 18F Fluorodeoxyglucose PET/CT Allow Prediction of Distant Metastasis. Radiology 2016;281:270-8. [Crossref] [PubMed]
- Kirienko M, Cozzi L, Rossi A, et al. Ability of FDG PET and CT radiomics features to differentiate between primary and metastatic lung lesions. Eur J Nucl Med Mol Imaging 2018;45:1649-60. [Crossref] [PubMed]
- Han H, Chen H. Selective lymph node dissection in early-stage non-small cell lung cancer. J Thorac Dis 2017;9:2102-7. [Crossref] [PubMed]
- Bustos García de Castro A, Ferreirós Domínguez J, Delgado Bolton R, et al. PET-CT in presurgical lymph node staging in non-small cell lung cancer: The importance of false-negative and false-positive findings. Radiología 2017;59:147-58. (English Edition). [PubMed]
- Li S, Zheng Q, Ma Y, et al. Implications of false negative and false positive diagnosis in lymph node staging of NSCLC by means of 18F-FDG PET/CT. PLoS One 2013;8:e78552. [Crossref] [PubMed]
- Cong M, Feng H, Ren J-L, et al. Development of a predictive radiomics model for lymph node metastases in pre-surgical CT-based stage IA non-small cell lung cancer. Lung Cancer 2020;139:73-9. [Crossref] [PubMed]
- Schimmel WCM, Gehring K, Eekers DBP, et al. Cognitive effects of stereotactic radiosurgery in adult patients with brain metastases: A systematic review. Adv Radiat Oncol 2018;3:568-81. [Crossref] [PubMed]
- Xu X, Huang L, Chen J, et al. Application of radiomics signature captured from pretreatment thoracic CT to predict brain metastases in stage III/IV ALK-positive non-small cell lung cancer patients. J Thorac Dis 2019;11:4516-28. [Crossref] [PubMed]
- Chen A, Lu L, Pu X, et al. CT-Based Radiomics Model for Predicting Brain Metastasis in Category T1 Lung Adenocarcinoma. AJR Am J Roentgenol 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Hosny A, Parmar C, Coroller TP, et al. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med 2018;15:e1002711. [Crossref] [PubMed]
- Wang T, Deng J, She Y, et al. Radiomics Signature Predicts the Recurrence-Free Survival in Stage I Non-Small-Cell Lung Cancer. Ann Thorac Surg 2020. [Epub ahead of print]. [Crossref] [PubMed]
- Wei H, Yang F, Liu Z, et al. Application of computed tomography-based radiomics signature analysis in the prediction of the response of small cell lung cancer patients to first-line chemotherapy. Exp Ther Med 2019;17:3621-9. [Crossref] [PubMed]
- Arshad MA, Thornton A, Lu H, et al. Discovery of pre-therapy 2-deoxy-2-18F-fluoro-D-glucose positron emission tomography-based radiomics classifiers of survival outcome in non-small-cell lung cancer patients. Eur J Nucl Med Mol 2019;46:455-66. [Crossref] [PubMed]
- van Timmeren JE, van Elmpt W, Leijenaar RTH, et al. Longitudinal radiomics of cone-beam CT images from non-small cell lung cancer patients: Evaluation of the added prognostic value for overall survival and locoregional recurrence. Radiother Oncol 2019;136:78-85. [Crossref] [PubMed]
- Konert T, Everitt S, La Fontaine MD, et al. Robust, independent and relevant prognostic 18F-fluorodeoxyglucose positron emission tomography radiomics features in non-small cell lung cancer: Are there any? PLos One 2020;15:e0228793. [Crossref] [PubMed]
- Bradley JD, Paulus R, Komaki R, et al. Standard-dose versus high-dose conformal radiotherapy with concurrent and consolidation carboplatin plus paclitaxel with or without cetuximab for patients with stage IIIA or IIIB non-small-cell lung cancer (RTOG 0617): a randomised, two-by-two factorial phase 3 study. Lancet Oncol 2015;16:187-99. [Crossref] [PubMed]
- Putora PM, Leskow P, McDonald F, et al. International guidelines on stage III N2 nonsmall cell lung cancer: surgery or radiotherapy? ERJ Open Res 2020;6:00159-2019. [Crossref] [PubMed]
- Coroller TP, Agrawal V, Huynh E, et al. Radiomic-Based Pathological Response Prediction from Primary Tumors and Lymph Nodes in NSCLC. J Thorac Oncol 2017;12:467-76. [Crossref] [PubMed]
- Yuan Y, Huang Q, Gu C, et al. Disease-free survival improved by use of adjuvant EGFR tyrosine kinase inhibitors in resectable non-small cell lung cancer: an updated meta-analysis. J Thorac Dis 2017;9:5314-21. [Crossref] [PubMed]
- Lu D, Wang Z, Liu X, et al. Differential effects of adjuvant EGFR tyrosine kinase inhibitors in patients with different stages of non-small-cell lung cancer after radical resection: an updated meta-analysis. Cancer Manag Res 2019;11:2677-90. [Crossref] [PubMed]
- David SE, Douglas EW, Charu A, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 1.2020. Journal of the National Comprehensive Cancer Network J Natl Compr Canc Netw 2019;17:1464-72. [Crossref] [PubMed]
- Wang F, Diao XY, Zhang X, et al. Identification of genetic alterations associated with primary resistance to EGFR-TKIs in advanced non-small-cell lung cancer patients with EGFR sensitive mutations. Cancer Commun (Lond) 2019;39:7. [Crossref] [PubMed]
- Zhu X, Chen L, Liu L, et al. EMT-Mediated Acquired EGFR-TKI Resistance in NSCLC: Mechanisms and Strategies. Front Oncol 2019;9:1044. [Crossref] [PubMed]
- Song J, Shi J, Dong D, et al. A New Approach to Predict Progression-free Survival in Stage IV EGFR-mutant NSCLC Patients with EGFR-TKI Therapy. Clin Cancer Res 2018;24:3583-92. [Crossref] [PubMed]
- Li H, Zhang R, Wang S, et al. CT-Based Radiomic Signature as a Prognostic Factor in Stage IV ALK-Positive Non-small-cell Lung Cancer Treated With TKI Crizotinib: A Proof-of-Concept Study. Front Oncol 2020;10:57. [Crossref] [PubMed]
- Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321-30. [Crossref] [PubMed]
- Oh SJ, Lee J, Kim Y, et al. Far Beyond Cancer Immunotherapy: Reversion of Multi-Malignant Phenotypes of Immunotherapeutic-Resistant Cancer by Targeting the NANOG Signaling Axis. Immune Netw 2020;20:e7. [Crossref] [PubMed]
- Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 2018;19:1180-91. [Crossref] [PubMed]
- Trebeschi S, Drago SG, Birkbak NJ, et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann Oncol 2019;30:998-1004. [Crossref] [PubMed]
- Khorrami M, Prasanna P, Gupta A, et al. Changes in CT Radiomic Features Associated with Lymphocyte Distribution Predict Overall Survival and Response to Immunotherapy in Non-Small Cell Lung Cancer. Cancer Immunol Res 2020;8:108-19. [Crossref] [PubMed]