
Peptidome characterization of ovarian cancer serum and the identification of tumor suppressive peptide ZYX36-58
Introduction
Ovarian cancer (OC) is the leading cause of gynecological cancer deaths. Due to the lack of early symptoms, physical signs, and effective early screening approaches, approximately 70% of OC cases are diagnosed at advanced stages, and the 5-year survival rate is less than 50% (1). Although great progress has been made in targeted therapy and immunotherapy of ovarian cancer during the past decade, overall survival has not significantly changed thus far (1). It therefore remains critical to discover new therapeutic targets and develop drugs with high-efficacy and low side effects for the treatment of ovarian cancer.
Due to the small molecular weight, short half-life, high targeting efficiency, low immunogenicity, peptides have been considered as promising drugs for the treatment of diseases (2). For example, ALM201, a therapeutic peptide derived from FKBPL, could disrupt angiogenesis and inhibit self-renewal of cancer stem cells and thus inhibit ovarian cancer growth, and has completed a Phase Ia clinical trial in ovarian cancer patients (3). The octapeptides LR and its more rigid diastereomer [d-Gln4]LR, could reduce the catalytic activity of human thymidylate synthase, and thus inhibit ovarian cancer growth (4). TAT-p + p-8 peptide (PPPPPPPPVDYEDEE) could efficiently disrupt the ABI1-EPS8 interaction, and block the invasion and metastasis of ovarian cancer cells (5). However, few studies have focused on endogenous peptides against ovarian cancer.
Indeed, endogenous peptides that exist naturally in vivo, such as oxytocin, thymosin, and insulin, are involved in development or reproduction, and are often clinically useful. Some endogenous peptides with anti-tumor properties have been reported (6-8) and even applied in clinic (9,10). With the development of liquid chromatography-tandem mass spectrometry (LC-MS/MS), more and more studies have focused on the endogenous peptides and their functions in vivo. Indeed, the identification of differentially expressed peptides in serum using LC-MS/MS has provided important clues for the diagnosis of various diseases, such as autism spectrum disorder of children (11), gestational diabetes mellitus (12), and colorectal cancer (13).
Interestingly, we noticed that several serum biomarkers are also regulators of cancer progression, such as miR-206 in cervical cancer (14,15), circPRMT5 in urothelial carcinoma (16). Besides, as important diagnostic biomarkers in ovarian cancer, CA125, which was also expressed in ovarian cancer cells can interact with mesothelin and thus contribute to the metastasis of ovarian cancer cell to the peritoneum (17), HE4 overexpression in ovarian cancer cells could promote ovarian cancer cell progression and metastasis in vitro and in vivo (18). These findings indicated that the differentially expressed endogenous peptides in serum may provide therapeutic targets for ovarian cancer treatment.
In this study, we first used LC-MS/MS to identify differentially expressed peptides in the serum of ovarian cancer patients and healthy controls, three peptides derived from the precursor proteins F13A1, ZYX and SNP23, respectively, which were down-regulated in the serum of ovarian cancer patients were chosen for the functional analysis. We found that only ZYX36-58 could significantly inhibit the invasion and migration of ovarian cancer cells. Therefore, we chose ZYX36-58 for further research. Our results show that the endogenous peptide ZYX36-58 has great promise in the treatment of ovarian cancer.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/atm-20-2018).
Methods
Sample collection
Serum from three epithelial ovarian cancer patients (48±25 years old, women) and three age-matched healthy controls (42±16 years old, women) were collected from Women’s Hospital of Nanjing Medical University (Nanjing Maternity and Child Health Care Hospital), and all experiments performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013), approved by the ethics committee of Women’s Hospital of Nanjing Medical University {Nanjing Maternity and Child Health Care Hospital, Ethics Approval Number: 2016 [16]} and all patients signed informed consent. Blood samples were centrifuged at 3,000 ×g for 10 min at 4 °C immediately after collection, then the serum supernatant was collected, supplemented with protease inhibitor (EDTA-free, Roche Applied Science, China), and quickly stored at −80 °C.
Peptide extraction
Peptide extraction and LC-MS/MS was performed as previously described (12). In brief, samples were thawed on ice, then centrifuged at 12,000× g for 15 min at 4 °C. The supernatants were collected and protein concentration was determined using the bicinchoninic acid kit (BCA, pierce, Rockford, IL, USA). Acetonitrile (ACN, 20% v/v) was then added to the samples and briefly vortexed, then incubated at room temperature for 30 min. Samples were then transferred to 10 kDa molecular weight cut-off centrifugal filter units (Millipore, USA), which was washed with 0.5 mL H2O before use. The peptides were extracted according to the manufacturer’s protocol of 10 kDa molecular weight cut-off centrifugal filter, then samples were dried in a SpeedVac (Scan Speed Maxi Vac, Labogene, Denmark). Tandem mass tag (TMT) labeling of the samples was conducted according to the manufacturer’s protocol (TMT 6-plex label reagent, Thermo Fisher Scientific, CA, USA). Finally, analysis was performed using nanoflow liquid chromatography coupled with LTQ-Qrbitrap Velos mass spectrometry.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS)
A 15 cm×75 µm i.d. (flow rate 300 nL/min) silica capillary column in series packed with 3.6 µm Jupiter C18-bonded particles was used for the analysis of the peptides in the gel. The separation was performed according to the effective gradient profiles, 5% mobile phase B (98% ACN, 0.1% FA) for 0–8 min, 8–35% mobile phase B for 8–43 min, 35–60% mobile phase B for 43–48 min, 60–80% mobile phase B for 48–50 min, 80% mobile phase B for 50–55 min, and 5% mobile phase B, 55–56 min. The separated peptide was then ionized by nanoESI and operated in the data-dependent acquisition mode by LTQ Orbitrap Velos (Thermo Fisher Scientific). The main parameters are as follows: voltage 1.8 V, scanning range of primary mass spectrometry 350≤1,500 m/z, resolution set to 30,000, starting m/z of secondary mass spectrometry fixed to 100, and resolution set to 7,500. The screening conditions of the parent ions for secondary fragmentation are as follows: the charge ≥2 and the first 12 parent ions with peak intensity of more than 1,000. The collision energy of higher energy collision conditions (HCD) is set to 35, and the fragment ions are detected in Orbitrap. Finally, the data are matched with theoretical mass spectrometric data obtained by the database simulation.
Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis of the protein precursors of the differentially expressed peptides
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the differentially expressed peptide precursor proteins was analyzed by Databases for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/).
Synthetic peptides
The following 3 differentially expressed peptides between the serum of ovarian cancer patients and healthy controls were synthesized by Shanghai Science Peptide Biological Technology Co., Ltd and dissolved in water or DMSO at the desired concentrations: ZYX36-58: VNPFRPGDSEPPPAPGAQRAQMG; F13A18-38: NNSNAAEDDLPTVELQGVVPR; SNP23123-131: TNGQLQQPT; and the scrambled peptide for ZYX36-58: PPNMAVADERPQQPRSFPGGAGP.
Culture of human ovarian cancer cell lines
The SKOV3 cell line (Homo sapiens, human, RRID: CVCL_0532) was purchased from Cell Bank of Chinese Academy of Sciences (Shanghai, China), and the HO8910 cell line (Homo sapiens, human, RRID: CVCL_6868) was purchased from Jiangsu Kaiji Biotechnolgy (Jiangsu, China). SKOV3 cells were cultured in McCoy’s 5A medium supplemented with 10% FBS, 2200 mg/L sodium bicarbonate and 2 mM L-glutamine, HO8910 cells were cultured in DMEM medium supplemented with 10% FBS. Both cell lines were maintained at 37 °C in a humidified incubator under 5% CO2.
Cell counting kit 8 assay
A suspension of about 1,000 cells and 100 µM ZYX36-58 peptide or peptide solvent in 100 µL culture medium were seeded in 96-well plate and incubated as above. Four hours after seeding, most cells were adherent to the surface of the plate, and this time point was considered as 0h. After 0, 12, 24 and 48 h of culture, the medium was supplemented with 10% CCK8 agent (Cell counting kit 8; Jiangsu Kaiji Biotechnolgy, Jiangsu, China) and continue to maintain at 37 °C in humidified incubator under 5% CO2 for 1 h. The absorbance at 450 nm was measured using a BioTek Synergy H4 instrument (BioTek Instruments, Vermont, USA).
Cell invasion and migration assay
About 800 µL medium containing 20% FBS was added to the lower chamber of a transwell (8-µm pore size, Millipore, Billerica, MA, USA) and 5×104 cells in 200 µL of medium supplemented with 100 µM ZYX36-58 peptide or peptide solvent were seeded in the upper chamber with or without Matrigel (Matrigel: medium =1:6, 60 µL/well) for invasion and migration assays, respectively, and incubated as above. After culturing for 24 h (for migration assay of SKOV3 cells) or 48 h (for invasion assay of SKOV3 cells, and for migration and invasion assay of HO8910 cells), the chamber was fixed in 4% paraformaldehyde for 60 min, followed by staining with 0.1% crystal violet for 30 min, washing two times in PBS, and removing the cells in the surface of the upper chamber by cotton swab. Three randomly selected areas of the lower surface of the chamber were selected for photography under Axio Observer D1 fluroscence microscope (Carl Zeiss, Germany). Finally, the cells on the lower surface were lysed in RIPA buffer, and the absorbance at 562 nm of the cell lysate was analyzed by the BioTek Synergy H4 instrument.
Fluorescence microscopy
SKOV3 cells were grown in 6-well plates, then FITC-labeled ZYX36-58 peptide was added to the culture medium at a concentration of 100 µM. Cell fluorescence was visualized and photographed at 1, 2, 4, 24 and 48 h after seeding under Axio Observer D1 fluroscence microscope (Carl Zeiss, Germany).
Peptide pull-down assay
Biotin-labeled ZYX36-58 or biotin-labeled scrambled peptide at a concentration of 100 µM was added to the medium of SKOV3 cells at 50% confluency, after 24 h culture, 1 ml of RIPA was added and total protein was extracted according to the manufacturer’s protocol. Then the total protein of each sample was incubated with 20 µL streptavidin-conjugated immunomagnetic beads overnight on a rotary shaker at 4 °C. The protein lysate containing the magnetic beads was placed in a Magna GrIP™ magnetic stand (Merck KGaA, Darmstadt, Germany) and the magnetic beads were washed by the protein lysate (RIPA: PMSF =100:1) 4–5 times. After 30 µL protein loading buffer (1×) was added to the magnetic beads, the mixture was heated in a 100 °C water bath for 5 min. Then the magnetic beads were discarded and the protein samples were used for sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After SDS-PAGE was completed, the gel was stained with silver according to the manufacturer’s protocol for Thermo Fisher Scientific Pierce silver stain kits, and the silver stained lane of ZYX36-58 and scrambled peptide treated group were separately excised for subsequent mass spectrometry analysis.
Protein lysis of the gel
Protein from the gel was extracted after washing, dehydration in acetonitrile, disulfide bond reduction in DTT, alkylation cysteine blocking by iodoacetamide, proteolysis, 50% acetonitrile extraction, and 100% acetonitrile extraction. After freeze-drying in the SpeedVac (Scan Speed Maxi Vac, Labogene, Denmark), the freeze-dried peptide powder was dissolved in mobile phase A (2% ACN, 0.1% FA), centrifuged at 20,000 ×g for 10 min, the peptides in the supernatant was separated by the Shimadzu LC-20AD liquid chromatograph system (Shimadzu, Kyoto, Japan).
Apoptosis assay
Cells were stained with PE and Annexin V using the PE-Annexin V Apoptosis Detection kit (BD Biosciences, San Diego, USA) after culturing with ZYX36-58 or peptide solvent and apoptosis inducer from the kit for 24 h. Cell apoptosis was analyzed with a flow cytometer (Beckman, CA, USA).
Wound healing assay
HO8910 and SKOV3 cells were cultured in 6-well plates in a medium containing 10% FBS and incubated for 24–36 h until the cell density reached 90–100%. The cells were maintained in serum-free medium for 6–8 h and a linear scratch was created on the cell layer using a 200 µL pipette tip, then the serum-free medium supplemented with peptide was added and replaced after 24 h of culture. The wound healing process was observed under EVOS XL Core digital microscope (Thermo Fisher Scientific, Waltham, USA) and analyzed using Image J software (Bethesda, USA).
Next-generation sequencing
About 1 mL TRIzol was added to the SKOV3 cells treated with ZYX36-58 or peptide solvent for 48 h. Three replicates of the TRIzol-treated cells in each group were used for next-generation sequencing. RNA was extracted according to the manufacturer’s protocol of TRIzol and RNA quality was assessed by Agilent 2100. The qualified RNA was then treated with the following procedures: fragmented after rRNA removal or mRNA enrichment, reverse transcribed by random N6 primer, synthesis of the double-stranded DNA and ER/A-tailing, ligating of the bubble adapter, PCR amplification using the specific primer, and then the library was constructed and sequenced by using the BGISEQ-500 system (BGI, Shenzhen, China).
Quantitative real-time PCR (qRT-PCR)
After culturing of cells with the ZYX36-58 or peptide solvent for 24 h, the cells were lysed in 1 mL TRIzol reagent, total RNA was isolated according to the manufacturer’s protocol, and cDNAs were synthesized using the First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). An aliquot of 20 ng of sample was subjected to qPCR analysis using the primers listed in Table S1. Raw data were analyzed according to the 2-ΔΔCT method.

Full table
Western blot
Total protein was extracted after lysing cells with RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) supplemented with protease inhibitor. The protein was then denatured by mixing with sodium dodecyl sulfonate (SDS) and boiled at 95 °C for 5 min. Next, an aliquot of 40 mg of protein sample was separated by 10% PAGE gel and immunoblots were visualized using electrochemiluminescence. The antibodies used in this study were anti-EEF1A2 (Proteintech, 16091-1-AP, RRID: AB_2096979), anti-YBX1 (Proteintech, 20339-1-AP, RRID: AB_10665424), anti-PCLO (Invitrogen, MA5-28620, RRID: AB_2745579), anti-TSP1 (Cell Signaling Technology, #37879, RRID: AB_2799123), anti-ZYX (Proteintech, 60254-1-AP, RRID: AB_2221279), anti-β-ACTIN (Santa Cruz, sc-47778, RRID: AB_626632), goat anti-mouse IgG (H+L) (Proteintech, SA00001-1, RRID: AB_2722565), and goat anti-rabbit IgG (H+L) (Millipore, AP132P, RRID: AB_92486).
Statistical analysis
Statistical analysis of the differentially expressed peptides used the two-tailed Students’ t-test, with P<0.05 and fold change ≥1.5 was considered statistically significant. The differentially expressed genes were analyzed by DEGseq software (19), with q<0.01 and fold change ≥1.5 was considered statistically significant. The proliferation, invasion, migration, apoptosis, qRT-PCR and WB were performed at least in triplicate and repeated at least three times with similar results. The pull down experiment was performed at least in duplicate. The data are presented as mean ± standard deviation in bar charts from at least three technical replicates, and the difference were analyzed by Student’s t-test, with P<0.05 considered statistically significant.
Results
The identification of the differentially expressed peptides in the serum of ovarian cancer patients and healthy controls
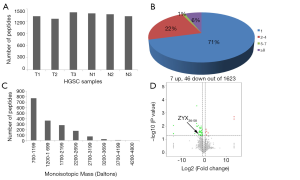
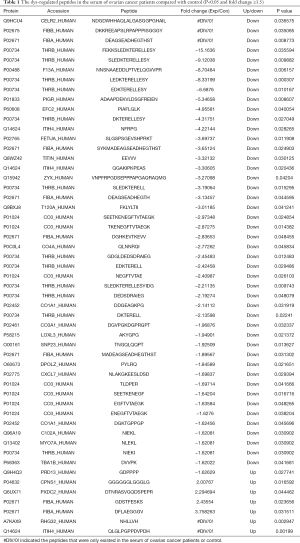
First, liquid chromatography-tandem mass spectrometry (LC-MS/MS) were performed for the identification of the differentially expressed peptides in the serum of ovarian cancer patients (n=3) and healthy controls (n=3). A total of 1,623 peptides derived from 495 precursor proteins were identified, with 1,331±80 peptides from 474 precursor proteins identified in control serum and 1,344±42 peptides from 461 precursor proteins identified in serum of ovarian cancer patients (Figure 1A and Table online: http://cdn.amegroups.cn/static/application/a02ebfdcb099287f46f5a43296c43bc5/atm-20-2018-1.pdf). Although 71% of the precursor proteins contained only 1 peptide, 22% of the precursor proteins contained 2–4 peptides, 1% of the precursor proteins contained 5–7 peptides, and 6% of the precursor proteins contained ≥8 peptides (Figure 1B). About 99.4% of the peptides were 5 to 26 amino acids in length and 98.6% of the peptides were 700 to 3,199 daltons in molecular weight (Figure 1C). In the serum of ovarian cancer patients, 7 peptides that were derived from 6 precursor proteins were up-regulated and 46 peptides that were derived from 23 precursor proteins were down-regulated (P<0.05 and fold change ≥1.5, Figure 1D and Table 1). We also noticed that two peptides from FIBA protein and 1 peptide from ITIH4 protein were up-regulated whereas 5 peptides from FIBA protein and 2 peptides from ITIH4 protein were down-regulated in the serum of the ovarian cancer patients (Table 1), indicating that these peptides have different half-life in different systems, so we propose that they may also perform biological functions and are strictly regulated.
Full table
Gene ontology and pathway analysis of the precursor proteins of the differentially expressed peptides
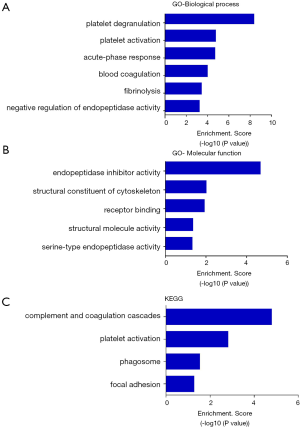
It was once thought that only intact proteins are functional units in the body. Then we found that the activation of a variety of proteins depends on proteolytic processes. As a product of the proteolytic products, endogenous peptides can also directly interact with target cells or target proteins and thus regulate the targets’ function (2). Indeed, most of the peptides’ function was closely related with their precursor proteins (2). So gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the precursor proteins of the differentially expressed peptides were performed. GO analysis indicated that the precursor proteins were mainly involved in the biological processes, such as platelet degranulation, platelet activation, the acute-phase response, blood coagulation, fibrinolysis and the negative regulation of endopeptidase activity (Figure 2A), involved in the molecular functions, such as endopeptidase inhibition, cytoskeletal structure, receptor binding, structural molecule activity and the serine-type endopeptidase activity (Figure 2B). KEGG pathway analysis indicated that the precursor proteins mainly involved in the complement and coagulation cascades, platelet activation, phagosome, and focal adhesion pathways (Figure 2C).
ZYX36-58 inhibits the invasion, migration and promotes apoptosis of ovarian cancer cells
Previous study has shown that the platelet activation and platelet-cancer cell interaction are critical factors for cancer metastasis (20), complement system activation in tumor microenvironment promotes tumor growth and metastasis (21), and several molecules in focal adhesion pathway are involved in ovarian cancer cell proliferation and migration (22). Besides, several coagulation factors contribute to cancer initiation, growth and metastasis (23) and the molecules essential for the phagosome are important regulators of autophagy and cancer progression (24). Besides, although none of the dys-regulated peptides have been reported to be the diagnostic biomarker, peptides (DGEA and DGEAGK) with the recognition motif DEGA from type I collagen were identified with the ability to bind α2β1-integrin, so cy5.5 labeled DEGA and DGEAGK was used for optical imaging in mice with α2β1 positive prostate tumors (25). Fibrinogen derived peptides alphastatin (ADSGEGDFLAEGGGVRGPRVVERH) and AHN419 (DFLAEGGGVRG) could inhibit endothelial cell tubule formation and migration in vitro and subcutaneous breast tumor growth in vivo (26). In our results, one of the down-regulated peptides from COL1A1 (DDGEAGKPG) contains the recognition motif, and one of the up-regulated peptide derived from FIBA protein (DFLAEGGGV) share the sequence DFLAEGGGV with alphastatin and AHN419 peptides. So we propose that some of the differentially expressed peptides may also function in the progression of ovarian cancer.
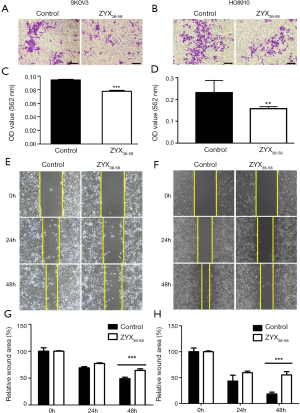
Three peptides which were down-regulated in ovarian cancer patients (ZYX36-58, F13A18-38, and SNP23123-131, detailed information of these peptides are presented in Table 1) were selected and synthesized. They were derived from the focal adhesion protein Zyxin (ZYX), Coagulation factor XIII A chain (F13A), Synaptosomal-Associated Protein 23 [SNP23, an important regulator of phagosome formation (27)], respectively. The cell proliferation, invasion, migration and apoptosis of cultured HO8910 and SKOV3 cells were analyzed after treatment with these peptides or peptide solvent. Our results showed that cell invasion and migration was significantly inhibited in HO8910 and SKOV3 cells treated with ZYX36-58 peptide (Figure 3) but not with other peptides (data not shown). Meanwhile, proliferation of HO8910 and SKOV3 cells were not altered after treatment with ZYX36-58 peptide (Figure 4A,B). But apoptosis of HO8910 and SKOV3 was significantly increased after treatment with ZYX36-58 peptide (Figure 4C,D,E,F). In conclusion, ZYX36-58 treatment in vitro can significantly inhibit cell invasion and migration, and promote cell apoptosis of ovarian cancer cells, but has no effect on ovarian cancer cell proliferation.
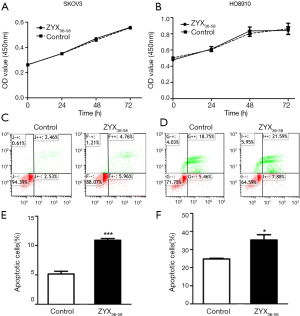
The mechanism of ZYX36-58 function on ovarian cancer cell invasion, migration and apoptosis
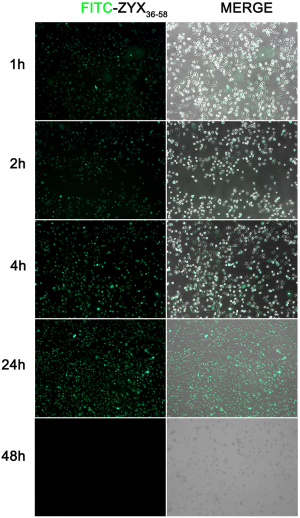
To explore the mechanism whereby ZYX36-58 inhibits invasion and migration of ovarian cancer cells, we began by analyzing the peptide distribution using the FITC-labeled ZYX36-58. The results showed that FITC-labeled ZYX36-58 was observed within 1 h in both the cell matrix and nucleus; and the fluorescence intensity peaked at 24 h, but was diminished by 48 h (Figure S1). Then we first analyzed the interaction between ZYX36-58 and its precursor protein ZYX by biotin-labeled ZYX peptide pull down and western blot, however, ZYX was not detected in the eluent of ZYX36-58 (Figure S2A). Then how does ZYX36-58 function in inhibiting cell invasion, migration and promoting cell apoptosis of ovarian cancer cells?
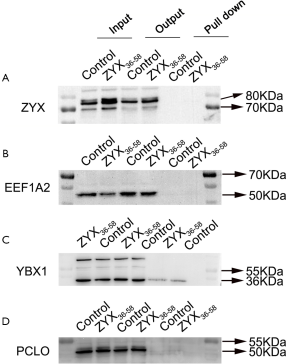
Mass spectrometry followed by biotin-labeled peptide pull down assay was an important strategy for identifying the peptide interacting proteins. The protein pull-down by biotin-labeled ZYX36-58 and biotin-labeled scrambled peptide were first analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining (Figure 5A). As a result, several protein bands were specifically found in biotin-labeled ZYX36-58 group. So the in-gel digested proteins from the band of biotin-labeled ZYX36-58 and biotin-labeled scrambled peptide as illustrated in Figure 5A were analyzed by LC-MS/MS. Finally, 22 proteins were specifically identified in the biotin-labeled ZYX36-58 pull down lysate (Table S2), and 68 proteins were specifically identified in the biotin-labeled scrambled peptide pull down lysate. Among the proteins specifically identified in the biotinylated ZYX36-58 group, EEF1A2, YBX1, PCLO, PROF1, RAP1L, TSP1, RACK1, A2MG and TLN1 were also involved in the invasion and migration of cancer cells. So further western blot analysis of the top four enriched proteins (EEF1A2, TSP1, YBX1 and PCLO) followed by biotin-labeled ZYX36-58 pull down assay was performed, our results showed that only physical interaction between TSP1 (Thrombospondin-1) and ZYX36-58 was detected, but no interaction was observed between ZYX36-58 and the other three proteins (Figure 5B and Figure S2B,C,D). Intriguingly, the protein level of TSP1 was also significantly increased in ZYX36-58 treated ovarian cancer cells (Figure 5C,D), which indicated that ZYX36-58 may function through binding and upregulating TSP1 protein level.
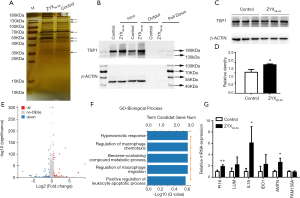

Full table
Next, the differentially expressed mRNA in ZYX36-58 treated and untreated ovarian cancer cells were analyzed by high throughput sequencing. Altogether, 169 genes were significantly upregulated and 49 genes were significantly down-regulated in ZYX36-58 treated ovarian cancer cells [fold change >1.5, FDR <0.01, Figure 5E and Table (online: http://cdn.amegroups.cn/static/application/42a7b771d63b6a426459e247cf00fdd4/atm-20-2018-2.pdf]. Gene ontology analysis of the biological pathway indicated that the differentially expressed genes are involved in the regulation of macrophage chemotaxis and macrophage migration, and the positive regulation of the leukocyte apoptotic process (Figure 5F). Further, qRT-PCR showed that gene expression of interleukin-1 alpha (IL1A), peptidase inhibitor 16 (PI16), and amelotin (AMTN), all of which are important inhibitors of cancer progression, was significantly up-regulated in ZYX36-58 treated ovarian cancer cells, which is consistent with the sequencing result (Figure 5G).
Interestingly, TSP1 exerts its antitumor effects through its widely established antiangiogenic ability (28), by inducing ovarian cancer apoptosis (6), via the regulation of immune system (29), and by activating TGF-β signaling pathway (30), and so on. Previous study indicated that thrombin can directly activate IL-1α (31) as well as de novo synthesis of TSP1 (32). Besides, PI16 is highly expressed in homing CD8+ T cells (33) and is considered as a CD4 subset biomarker (34). The secreted protein AMTN is induced at the initiation of apoptosis by TGF-β1 (35). These results indicated that ZYX36-58 may inhibit ovarian cancer invasion and migration through targeting TSP1 and its downstream signaling pathway.
Discussion
With the development of mass spectrometry, several studies have focused on the identification of the peptide biomarkers in diseases (11-13). Up to now, the peptidome of the urine from healthy control and ovarian cancer patients and the peptidome of the ascites from benign gynecological conditions and ovarian cancer patients have been analyzed (36,37), the serum peptidomes of control and ovarian cancer patients have not been studied before. However, none of the differentially expressed peptides in this study were found in the preceding studies. This could be caused by the different systems in different studies and may further reflect the peptide distribution specificity in different systems.
In this study, KEGG pathway analysis of the differentially expressed peptides indicated that although most of the peptides’ precursors are important regulators of the coagulation system, there are also some regulators of focal adhesion, platelet activation and phagosome formation, which are important for cancer initiation and progression (21,38). Besides, two of the dys-regulated peptides in our study also share the same sequence with the functional peptides reported before (24,25), so we accordingly think that some of the differentially expressed peptides are involved in ovarian cancer progression.
Endogenous peptides are increasingly seen to be important regulators of biological processes. For example, the production of endogenous peptide apelin, induced by muscle contraction, can significantly enhance muscle function by inducing the formation of mitochondria in muscle fibers and influencing autophagy and anti-inflammatory pathways (39). The peptide PAF, released by diverse therapeutic sensitive cancer cells following therapy, can promote apoptosis of therapy-resistant cancer cells (8). Serum A-type natriuretic peptide (ANP) in the circulatory system can significantly inhibit the proliferation of cancer cells in vitro and inhibit the cancer growth in vivo (40). Additionally, the C-type natriuretic peptide (CNP) and its precursor are considered as biomarkers of human prostate cancer (41), and CNP can also inhibit the cell proliferation of prostate cancer cells by inhibiting DNA synthesis (42). For this reason, we synthesized three peptides that were differentially expressed in serum of control vs. ovarian cancer patients and analyzed the function of these peptides on proliferation, invasion and migration of ovarian cancer cells. Our results clearly indicated that one of these peptides, ZYX36-58, can inhibit the invasion and migration of ovarian cancer cells and thus may become an important target of ovarian cancer therapy.
TSP1 is an extracellular matrix protein that can bind fibronectin, collagen and integrin, mediate cell adhesion and migration (43), bind VEGF and regulate the internalization and expression of VEGF and promotes the apoptosis of endothelial cells (28). A higher plasma TSP1 level predicts longer overall survival of ovarian cancer patients (44). TSP1 derived peptides such as ABT-510, ABT-526, DI-TSP, which mimic the thrombospondin-1/CD47 epitope, and the prosaposin-derived peptide targeting TSP1, all can inhibit the growth and metastasis of ovarian cancer and other types of cancer (6,45-47). This has caused secreted TSP1 to be considered an important therapeutic target of ovarian cancer. Interestingly, our results show that ZYX36-58 can bind and increase the protein level of TSP1.
TSP1, also known as the thrombin-sensitive protein, was first separated from thrombin-induced platelets (48). TSP1 regulates macrophage recruitment and the production of inflammatory factors such as IL-1β (49), and IL-10 (50). Gene ontology showed that the differentially expressed genes between ZYX36-58 and peptide solvent treated ovarian cancer cells are involved in the regulation of macrophage chemotaxis, macrophage migration, and leukocyte apoptosis, which is consistent with the peptide pull-down result. IL-1A, which was also upregulated in the ZYX36-58 treated ovarian cancer cells, whose activation directly links the coagulation system and immune system; this is also consistent with KEGG analysis of the precursor proteins of the differentially expressed peptides in the ZYX36-58 and peptide solvent treated ovarian cancer cells. Thus, although the mechanism by which ZYX36-58 increases TSP1 protein level is still unknown and the binding efficiency is low, our study provides a new strategy for identifying the therapeutic molecules.
In future, strategies for increasing the stability and target efficiency of ZYX36-58, such as N-terminal acetylation, C-terminal amination, cyclization and amino acid substitution, may also be used to increase its stability or ability and thereby improve the feasibility of the clinical application of ZYX36-58.
Conclusions
With small molecular weight, short half-life and low immunogenicity, peptides drugs are considered as one of the development directions with wide market prospect in the pharmaceutical industry. Indeed, several peptide drugs have been used in clinic, such as thymic pentapeptide (the peptide derived from thymopoietin II), Leuprorelin (GnRH analogue) and octreotide (Somatostatin octapeptide). With the development of mass spectrometry and peptide synthesis technology, more and more endogenous peptides have been identified, although only few of the peptide’s function have been characterized, we believe that most of the bioactive peptides’ function will be comprehensively analyzed, and the peptide drugs will be used in clinic in the future.
Acknowledgments
Funding: The study was financially supported by the National Natural Science Foundation of China (81602285, 81872126), Jiangsu provincial key research and development program (BE2019621), Nanjing Medical Science and Technique Development Foundation (Grant numbers: JQX17009, ZDX16015, ZKX15046, QRX17159), Research Innovation Program for Graduates of Jiangsu Province (JX10413664, JX22013553).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/atm-20-2018
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-2018
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-2018
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2018). XW, GL, XP, XJ and JX report a patent of a polypeptide ZYX36-58 for the treatment or adjuvant treatment of ovarian cancer pending. JX serves as an unpaid Section Editor of Annual of Translational Medicine from Oct 2019 to Mar 2020. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All experiments performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013), approved by the ethics committee of Women’s Hospital of Nanjing Medical University (Nanjing Maternity and Child Health Care Hospital, Ethics Approval Number: 2016[16]) and all patients signed informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Pan X, Xu J, Jia X. Research Progress Evaluating the Function and Mechanism of Anti-Tumor Peptides. Cancer Manag Res 2020;12:397-409. [Crossref] [PubMed]
- Annett S, Moore G, Short A, et al. FKBPL-based peptide, ALM201, targets angiogenesis and cancer stem cells in ovarian cancer. Br J Cancer 2020;122:361-71. [Crossref] [PubMed]
- Pelà M, Saxena P, Luciani R, et al. Optimization of peptides that target human thymidylate synthase to inhibit ovarian cancer cell growth. J Med Chem 2014;57:1355-67. [Crossref] [PubMed]
- Yu X, Liang C, Zhang Y, et al. Inhibitory short peptides targeting EPS8/ABI1/SOS1 tri-complex suppress invasion and metastasis of ovarian cancer cells. BMC Cancer 2019;19:878. [Crossref] [PubMed]
- Wang S, Blois A, El RT, et al. Development of a prosaposin-derived therapeutic cyclic peptide that targets ovarian cancer via the tumor microenvironment. Sci Transl Med 2016;8:329ra34. [Crossref] [PubMed]
- Soragni A, Janzen DM, Johnson LM, et al. A Designed Inhibitor of p53 Aggregation Rescues p53 Tumor Suppression in Ovarian Carcinomas. Cancer Cell 2016;29:90-103. [Crossref] [PubMed]
- Hebbar N, Burikhanov R, Shukla N, et al. A Naturally Generated Decoy of the Prostate Apoptosis Response-4 Protein Overcomes Therapy Resistance in Tumors. Cancer Res 2017;77:4039-50. [Crossref] [PubMed]
- Im SA, Lu YS, Bardia A, et al. Overall Survival with Ribociclib plus Endocrine Therapy in Breast Cancer. N Engl J Med 2019;381:307-16. [Crossref] [PubMed]
- Han W, Youn HJ. Clinical Studies Investigating the Use of Leuprorelin in Breast Cancer Patients from Asia. Asian Pac J Cancer Prev 2019;20:1475-9. [Crossref] [PubMed]
- Yang J, Chen Y, Xiong X, et al. Peptidome Analysis Reveals Novel Serum Biomarkers for Children with Autism Spectrum Disorder in China. Proteomics Clin Appl 2018;12:e1700164. [Crossref] [PubMed]
- Yin L, Huai Y, Zhao C, et al. Early Second-Trimester Peptidomic Identification of Serum Peptides for Potential Prediction of Gestational Diabetes Mellitus. Cell Physiol Biochem 2018;51:1264-75. [Crossref] [PubMed]
- Wang H, Luo C, Zhu S, et al. Serum peptidome profiling for the diagnosis of colorectal cancer: discovery and validation in two independent cohorts. Oncotarget 2017;8:59376-86. [Crossref] [PubMed]
- Wang Y, Tian Y. miR-206 Inhibits Cell Proliferation, Migration, and Invasion by Targeting BAG3 in Human Cervical Cancer. Oncol Res 2018;26:923-31. [Crossref] [PubMed]
- Chen AH, Qin YE, Tang WF, et al. MiR-34a and miR-206 act as novel prognostic and therapy biomarkers in cervical cancer. Cancer Cell Int 2017;17:63. [Crossref] [PubMed]
- Chen X, Chen RX, Wei WS, et al. PRMT5 Circular RNA Promotes Metastasis of Urothelial Carcinoma of the Bladder through Sponging miR-30c to Induce Epithelial-Mesenchymal Transition. Clin Cancer Res 2018;24:6319-30. [Crossref] [PubMed]
- Rump A, Morikawa Y, Tanaka M, et al. Binding of ovarian cancer antigen CA125/MUC16 to mesothelin mediates cell adhesion. J Biol Chem 2004;279:9190-8. [Crossref] [PubMed]
- Lu R, Sun X, Xiao R, et al. Human epididymis protein 4 (HE4) plays a key role in ovarian cancer cell adhesion and motility. Biochem Biophys Res Commun 2012;419:274-80. [Crossref] [PubMed]
- Wang L, Feng Z, Wang X, et al. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010;26:136-8. [Crossref] [PubMed]
- Wang S, Li Z, Xu R. Human Cancer and Platelet Interaction, a Potential Therapeutic Target. Int J Mol Sci 2018;19:E1246. [Crossref] [PubMed]
- Reis ES, Mastellos DC, Ricklin D, et al. Complement in cancer: untangling an intricate relationship. Nat Rev Immunol 2018;18:5-18. [Crossref] [PubMed]
- Zhao X, Guan JL. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv Drug Deliv Rev 2011;63:610-5. [Crossref] [PubMed]
- Falanga A, Marchetti M, Vignoli A. Coagulation and cancer: biological and clinical aspects. J Thromb Haemost 2013;11:223-33. [Crossref] [PubMed]
- Mancias JD, Kimmelman AC. Mechanisms of Selective Autophagy in Normal Physiology and Cancer. J Mol Biol 2016;428:1659-80. [Crossref] [PubMed]
- Leung K. Cy5.5-Asp-Gly-Glu-Ala-Gly-Lys-(Arg)8-OH. 2012 Jan 6 [Updated 2012 Apr 5]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004-2013.
- Staton CA, Stribbling SM, Garcia-Echeverria C, et al. Identification of key residues involved in mediating the in vivo anti-tumor/anti-endothelial activity of Alphastatin. J Thromb Haemost 2007;5:846-54. [Crossref] [PubMed]
- Sakurai C, Hashimoto H, Nakanishi H, et al. SNAP-23 regulates phagosome formation and maturation in macrophages. Mol Biol Cell 2012;23:4849-63. [Crossref] [PubMed]
- Vikhanskaya F, Bani MR, Borsotti P, et al. p73 Overexpression increases VEGF and reduces thrombospondin-1 production: implications for tumor angiogenesis. Oncogene 2001;20:7293-300. [Crossref] [PubMed]
- Hayat SM, Bianconi V, Pirro M, et al. CD47: role in the immune system and application to cancer therapy. Cell Oncol (Dordr) 2020;43:19-30. [Crossref] [PubMed]
- Qin H, Qu C, Yamaza T, et al. Ossifying fibroma tumor stem cells are maintained by epigenetic regulation of a TSP1/TGF-beta/SMAD3 autocrine loop. Cell Stem Cell 2013;13:577-89. [Crossref] [PubMed]
- Burzynski LC, Humphry M, Pyrillou K, et al. The Coagulation and Immune Systems Are Directly Linked through the Activation of Interleukin-1alpha by Thrombin. Immunity 2019;50:1033-42.e6. [Crossref] [PubMed]
- Miao X, Rahman MF, Jiang L, et al. Thrombin-reduced miR-27b attenuates platelet angiogenic activities in vitro via enhancing platelet synthesis of anti-angiogenic thrombospondin-1. J Thromb Haemost 2018;16:791-801. [Crossref] [PubMed]
- Lupsa N, Ersek B, Horvath A, et al. Skin-homing CD8(+) T cells preferentially express GPI-anchored peptidase inhibitor 16, an inhibitor of cathepsin K. Eur J Immunol 2018;48:1944-57. [Crossref] [PubMed]
- Hope CM, Welch J, Mohandas A, et al. Peptidase inhibitor 16 identifies a human regulatory T-cell subset with reduced FOXP3 expression over the first year of recent onset type 1 diabetes. Eur J Immunol 2019;49:1235-50. [PubMed]
- Nakayama Y, Matsui S, Noda K, et al. TGFbeta1-induced Amelotin gene expression is downregulated by Bax expression in mouse gingival epithelial cells. J Oral Sci 2018;60:232-41. [Crossref] [PubMed]
- Huang X, Zhou J, Tang R, et al. Potential Significance of Peptidome in Human Ovarian Cancer for Patients With Ascites. Int J Gynecol Cancer 2018;28:355-62. [Crossref] [PubMed]
- Smith CR, Batruch I, Bauca JM, et al. Deciphering the peptidome of urine from ovarian cancer patients and healthy controls. Clin Proteomics 2014;11:23. [Crossref] [PubMed]
- Leung CS, Yeung TL, Yip KP, et al. Calcium-dependent FAK/CREB/TNNC1 signalling mediates the effect of stromal MFAP5 on ovarian cancer metastatic potential. Nat Commun 2014;5:5092. [Crossref] [PubMed]
- Vinel C, Lukjanenko L, Batut A, et al. The exerkine apelin reverses age-associated sarcopenia. Nat Med 2018;24:1360-71. [Crossref] [PubMed]
- Vesely BA, Fitz SR, Gower WJ, et al. Vessel dilator: most potent of the atrial natriuretic peptides in decreasing the number and DNA synthesis of human squamous lung cancer cells. Cancer Lett 2006;233:226-31. [Crossref] [PubMed]
- Lippert S, Iversen P, Brasso K, et al. C-type natriuretic peptide and its precursor: potential markers in human prostate cancer. Biomark Med 2015;9:319-26. [Crossref] [PubMed]
- Vesely BA, Alli AA, Song SJ, et al. Four peptide hormones' specific decrease (up to 97%) of human prostate carcinoma cells. Eur J Clin Invest 2005;35:700-10. [Crossref] [PubMed]
- Rosini S, Pugh N, Bonna AM, et al. Thrombospondin-1 promotes matrix homeostasis by interacting with collagen and lysyl oxidase precursors and collagen cross-linking sites. Sci Signal 2018;11:eaar2566.
- Cymbaluk-Płoska A, Chudecka-Glaz A, Pius-Sadowska E, et al. Thrombospondin-I concentrations behavior in plasma of patients with ovarian cancer. Cancer Biomark 2017;20:31-9. [Crossref] [PubMed]
- Greenaway J, Henkin J, Lawler J, et al. ABT-510 induces tumor cell apoptosis and inhibits ovarian tumor growth in an orthotopic, syngeneic model of epithelial ovarian cancer. Mol Cancer Ther 2009;8:64-74. [Crossref] [PubMed]
- Denèfle T, Pramil E, Gomez-Morales L, et al. Homotrimerization Approach in the Design of Thrombospondin-1 Mimetic Peptides with Improved Potency in Triggering Regulated Cell Death of Cancer Cells. J Med Chem 2019;62:7656-68. [Crossref] [PubMed]
- Rusk A, McKeegan E, Haviv F, et al. Preclinical evaluation of antiangiogenic thrombospondin-1 peptide mimetics, ABT-526 and ABT-510, in companion dogs with naturally occurring cancers. Clin Cancer Res 2006;12:7444-55. [Crossref] [PubMed]
- Ganguly P. Isolation and properties of a thrombin-sensitive protein from human blood platelets. J Biol Chem 1971;246:4286-90. [PubMed]
- Stein EV, Miller TW, Ivins-O'Keefe K, et al. Secreted Thrombospondin-1 Regulates Macrophage Interleukin-1beta Production and Activation through CD47. Sci Rep 2016;6:19684. [Crossref] [PubMed]
- Zhao Y, Xiong Z, Lechner EJ, et al. Thrombospondin-1 triggers macrophage IL-10 production and promotes resolution of experimental lung injury. Mucosal Immunol 2014;7:440-8. [Crossref] [PubMed]


