
Prognostic values of SNAI family members in breast cancer patients
Introduction
Breast cancer (BC) is one of the most frequently diagnosed cancers worldwide and is the leading cause of cancer-related death in females (1). China once had a relatively low incidence of BC, but since the 1990s, the growth rate has increased to twice that of the global rate, with BC being especially prevalent in urban areas (2). Although early diagnostic techniques have reached relative maturity, inherent barriers may contribute to a more frequent presentation of advanced-stage BC, especially in China (3,4), with the greater delay in diagnosis always resulting in worse prognosis and stage progression (5). Given these facts, the ability to predict the prognosis of BC by identifying potential prognostic markers is a crucial clinical need.
Snail family members of vertebrates, which include Snail1 (encoded by the SNAI1 gene), Snail2 (encoded by the SNAI2 gene and also named Slug), and Snail3 (encoded by the SNAI3 gene and previously named Smuc), are zinc finger proteins that function as transcriptional repressors (6). Given their nature, SNAI1 and SNAI2 have been extensively studied in humans for nearly a decade. Their main mechanism of effect was revealed to be a suppression of CDH1 transcription, which leads to a loss of the E-cadherin expression protein and the inducement of epithelial-to-mesenchymal transition (EMT) (7,8). It is well known that, besides participating in normal developmental processes, EMT plays a critical role in malignant transformation and tumor progression (9). Thus, it is widely accepted that SNAI1 and SNAI2 are associated with the malignant biological properties of cancer cells. Clinical data indicate that the reactivation of either of these two proteins is associated with a high rate of metastasis and a rather poor prognosis in various malignant tumors, including gastric cancer and lung cancer (10,11). SNAI3 was the most recent member of this family to be discovered, and unlike the other two proteins, less information about the function of SNAI3 is known. This protein shares the structure of the C-terminal DNA-binding domain and N-terminal transrepression domain with both of its fellow Snail members, SNAI1 and SNAI2.
Further research, based on analyzing the expression, location, and biological processes of the Snail family, has revealed other non-EMT–related functions (12,13). Several studies have demonstrated a correlation of Snail family with prognosis in various malignant tumors. A Chinese gastric cancer cohort reported that SNAI1 protein expression was closely linked to both clinicopathological characteristics and survival (P<0.001) (14). In BC, high SNAI1 expression detected by immunohistochemistry (IHC) in tumor tissue was found to be directly correlated with poor survival in breast invasive ductal carcinoma patients, regardless of human epidermal growth factor receptor 2 (ERBB2) status (15,16). The role of SNAI2 in various cancer types has also been investigated, with its protein expression being associated with unfavorable prognosis (17-19). However, there are limited reports concerning the prognostic value of SNAI3, and the few published studies that do exist are limited by the subjective interpretation of IHC methods or small sample sizes.
Therefore, the current study endeavored to evaluate the expression of Snail family members in a large cohort of BC patients taken from public databases. We here present the evidence drawn from this study, indicating a correlation between the expression of all three SNAI mRNAs with prognosis in BC patients.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/atm-20-681).
Methods
Based on the data from cBioPortal for Cancer Genomics (http://www.cbioportal.org/), we analyzed the genomic alterations of the three members of the SNAI family in solid tumors, as well as the co-expressed genes with these three genes in BC, respectively. After screening the co-expressed genes (Pearson’s correlation of >0.3 or <−0.3), the potential biological pathways and biological processes were further predicted with FunRich software (version 3.1) (20). Normalized mRNA expression profile data were downloaded from Gene Expression Omnibus (GEO) (http://bioprofiling.de/GEO/PPISURV/ppisurv.html). These data were used to analyze the prognostic value of these three SNAI family members in solid tumors. For further validation, we obtained the individual SNAI family members’ mRNA expressions in BC from UCSC Xena Browser (https://xenabrowser.net/). Clinical data, including intrinsic subtype, grade, estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor-2 (HER2) status, were collected. With the aim of assessing the prognostic values of the three Snail family members in BC, each member was entered into the database (http://kmplot.com/analysis/index.php?P=service&cancer=breast). The primary outcome was distant metastasis-free survival (DMFS), while recurrence-free survival (RFS) and overall survival (OS) were the secondary evaluation indicators of prognosis. DMFS, RFS, and OS were defined as latency to the first distant metastasis, disease relapse, and death, respectively. We then further analyzed the impact of the three individual Snail family members on the survival of BC patients with diverse clinicopathological characteristics. Ethical approval was not required because the data used in this study are from public databases, and informed consent was waived because of the retrospective nature of this study.
Kaplan-Meier plots with number-at-risk, 95% confidence intervals, log-rank P value, and hazard ratio (HR) were obtained from the webpage (http://kmplot.com/analysis/index.php?P=service&cancer=breast). Data from the Human Protein Atlas database (http://www.proteinatlas.org/) were utilized to analyze the expression level of individual Snail family members in solid tumors. A P-value of less than 0.05 was regarded as statistically significant.
Results
Differences in expression and gene alterations of individual Snail family members
The protein expression status of the Snail family members was evaluated in the HPA database. These three members showed different expressions across various normal tissues. SNAI1 protein levels were significantly high in gallbladder tissue, while those of SNAI2 and SNAI3 were relatively high in cervix and skeletal muscle, respectively. Normal breast tissue exhibited relatively low-level expressions of SNAI1 and SNAI3, whereas SNAI2 presented a markedly high level of expression (Figure S1A).
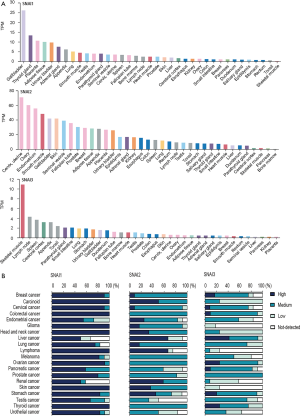
In tumor tissue, we discovered that SNAI1 was overexpressed among most of the malignant tumors. The expression of SNAI2 and SNAI3 in various tumors was frequently low (Figure S1B). In BC, nearly 90% of samples exhibited an overexpression of SNAI1, while the expression of SNAI2 and SNAI3 was often medium or low.
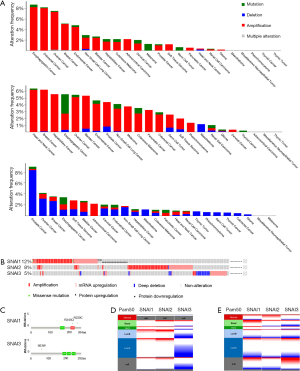
We also analyzed the genomic alternations in solid tumors using 11,165 samples from the cBioportal database. The most common alteration types were copy number amplification (CNA) in the SNAI1 and SNAI2 genes and copy number deletion in the SNAI3 gene (Figure 1A). In BC, 5% of patients presented SNAI1 alterations, with the most common being copy number alteration (CNA), followed by mRNA upregulation, protein downregulation, protein upregulation, and missense mutation (Figure 1B). The distribution of SNAI2 alterations was similar with that of SNAI1. However, only 5% of BC patients showed genetic SNAI3 alterations. mRNA upregulation and deep deletion were the two most common alternation types (Figure 1B). Furthermore, extremely low mutation frequency was present in BC, with the most frequently mutations being R243Q and R220C in the SNAI1 gene, and S63W in the SNAI3 gene, (Figure 1C). No SNAI2 mutations were observed in BC. The CNA and mRNA expression levels of the three SNAI genes are presented, based on the PAM50 subtype, in Figures 1D and 1E, respectively. SNAI1 and SNAI2 tended to show gene amplification in all subtypes, whereas, for SNAI3, deletion was predominant. For mRNA expression, the basal subtype mainly showed SNAI1 and SNAI2 upregulation, along with SNAI3 downregulation. A similar result was discovered in the HER2-enriched subtype. However, in the luminal A and luminal B subtypes, all three proteins were mainly downregulated, which contrasted starkly with the expressions found in the basal and HER2-enriched subtypes.
Prognostic values of Snail family members in BC patients
First, the prognostic value of the three Snail family members in solid tumors was explored using the data from Gene Expression Omnibus (GEO) (Supplementary 1, available at https://cdn.amegroups.cn/static/application/cce367de5ef80c1cb77dbb6c03be5009/10.21037atm-20-681-1.pdf). The survival effect produced by SNAI1 in all included tumors was significantly negative, while SNAI3 produced a positive effect on survival for these tumors. SNAI2 demonstrated neither a consistently negative nor positive impact on survival in different tumors. Meanwhile, high mRNA levels of SNAI1 mainly correlated with significantly poor survival in BC patients with premenopause status (P=0.0025) or triple-negative subtype (P=0.0285). In contrast, SNAI3 showed a positive relation between high expression and better survival in certain BC types, including lymph node–positive, progesterone receptor–negative, estrogen receptor–negative, and HER2 overexpression subtypes, and in those patients who received radiation treatment (Supplementary 1, available at https://cdn.amegroups.cn/static/application/cce367de5ef80c1cb77dbb6c03be5009/10.21037atm-20-681-1.pdf).
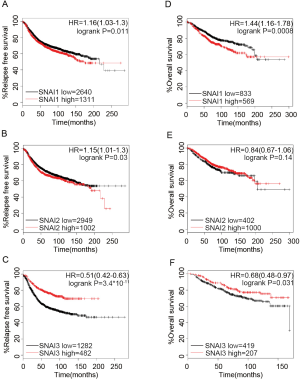
To validate the prognosis of the Snail family members in BC, we downloaded the data of BC patients from cBioPortal. In total, 1746 patients were detected with SNAI1 and SNAI2 expression, and 664 were detected with SNAI3. The Kaplan-Meier Plotter platform was further used to determine the association between the survival of BC patients and the expression of SNAI family members. The DMFS survival curves are shown in Figure 2. We observed that low SNAI1 mRNA expression was associated with better DMFS (Figure 2A, HR =1.55, 95% CI: 1.28–1.89, P=1.1×10-5). On the contrary, high mRNA expression of SNAI3 was significantly associated with favorable DMFS (Figure 2C, HR =0.49, 95% CI: 0.33–0.72, P=0.00026). For SNAI2, a slight statistical difference was observed for the effect of mRNA expression on prognosis (Figure 2B, HR =1.24, 95% CI: 1.01–1.53, P=0.042). Moreover, we confirmed that the Snail mRNA expression level was correlated with RFS and OS (Figure S2). The mRNA expression of any member of the SNAI family presented significant statistical difference, except for the effect of SNAI2 mRNA expression on OS.

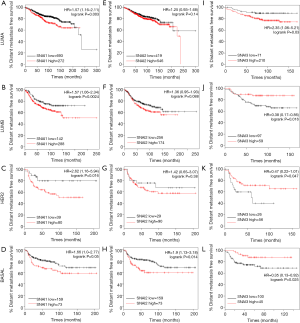
Additionally, we further explored whether Snail mRNA expression affected prognosis in BC with various PAM50 intrinsic subtypes (Figure 3). In all the four subtypes, SNAI1 was significantly associated with worse DMFS (Figure 3A,B,C,D), but SNAI3 was significantly correlated with better DMFS (Figure 3I,J,K,L). The significant correlation between SNAI2 and DMFS was only observed in the basal-like BC type (Figure 3H, P=0.014), and no association was found in the other types.
Prognostic values of SNAI family members in BC patients with various clinicopathological features
Next, we identified the prognostic values of the Snail expression of patients with different clinicopathological features, including grade, ER, PR, and HER2 status (Table 1). Regardless of grade II/III, ER, PR, lymph node status, or treatment history, high SNAI1 mRNA expression was associated with poor DMFS. For the TP53 wild-type patients, a correlation between SNAI1 and short DMFS was discovered, while no association was observed between SNAI1 expression and DMFS in TP53-mutant patients. The effect of SNAI2 expression on survival was dynamically based on different clinicopathological statuses, including ER, grade, TP53, and treatment history. Furthermore, high SNAI3 mRNA expression was associated with better DMFS in patients with any of TP53 status, the basal-like 2 subtype, and those who had undergone endocrine therapy or chemotherapy. The analyses of RFS and OS are shown in the Supplementary 2, available at https://cdn.amegroups.cn/static/application/8b93f873767d8d06f76b154eb3e22088/10.21037atm-20-681-2.pdf. For patients with most of the listed features, high SNAI1 expression was correlated with poor RFS and OS, while the opposite was true for high SNAI3 expression. Furthermore, the impact of SNAI2 expression on long-term survival varied with the diverse clinical features of patients.
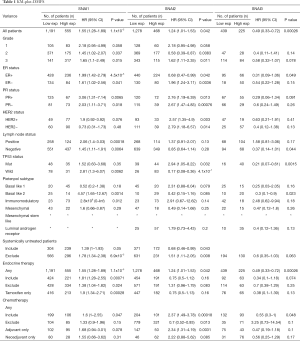
Full table
Pathway enrichment and biological process analysis of individual Snail family members and their co-expressed genes
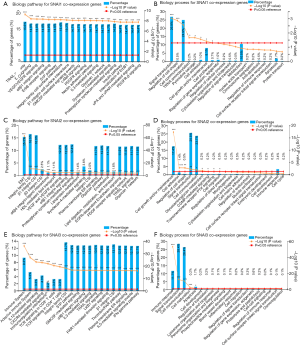
Lastly, the pathways and biological processes of the three different Snail family genes and their co-expressed genes were investigated (Supplementary 3, available at https://cdn.amegroups.cn/static/application/e19ccde1fe181a3fff3c28d98075596a/10.21037atm-20-681-3.pdf). The network reflected interactions between Snail family members and a total of 2053 non-redundant co-expressed genes. As shown in Figure 4A,B, SNAI1 was found to participate in the mediation of multiple biological processes including signal transduction, regulation of cell shape, and cell communication, and to carry out biological functions through multiple signaling pathways including the TRAIL pathway, ER signaling, VEGF/EGFR signaling, α9β1 integrin signaling, and others. SNAI2 and its co-expressed genes mainly enriched similar biological processes, including signal transduction and cell communication, and the close pathways, like integrin family cell surface interactions (CSI) (Figure 4C,D). For SNAI3 and its co-expressed genes, while most of the mapped processes and pathways were comparable with those of its two family members, immune-related items were also significantly enriched (Figure 4E,F).
Discussion
In the current study, we probed the public databases to determine the associations of Snail family members with survival outcomes in patients with BC. The results indicated that SNAI1 had a relatively higher expression in BC tissue than in most types of tumors but a lower expression in normal breast tissue, with its high expression correlating with worse survival in BC patients. The expression status of SNAI3 mRNA was different from that of SNAI1 in tumor and normal tissues and had the opposite impact on survival. SNAI3 might thus be a better potential prognostic biomarker in patients with BC. For patients with diverse clinicopathological features, a high expression of SNAI1 generally indicated worse DMFS. SNAI3 had a markedly higher expression in BC tissues, which mostly indicated favorable survival. However, the prognostic value of SNAI2 expression could not be determined. The current work is the first to report the expression of SNAI family members in BC by utilizing the public database.
The details concerning the function of SNAI1 on EMT have been reported, with SNAI1 showing a closer link with E-cadherin than the other two proteins (8). It is also a candidate prognostic factor in diverse malignant tumors, especially BC. In the present study, E-cadherin was not discovered in co-expressed genes of SNAI1, and the high expression of SNAI1 mRNA was still associated with poor survival. Our data confirmed that SNAI1 was a biomarker for BC prognosis again. Meanwhile, we found that SNAI3, the lesser-known Snail member, had a correlation with positive prognosis in BC. Preclinical research has indicated that decreased SNAI3 expression is associated with the increased expression of CDH1 via knockdown of FOXD4 (21), while a higher expression of CDH1 is significantly related with cell migration and invasion ability, which may lead to poor prognosis. In ovarian cancer, patients with SNAI3 high expression were found to have longer overall survival than those with low expression, demonstrating that SNAI3 is a good prognostic factor (HR =0.61, P=5.73×10-5) (22). These findings are consistent with our results and provide a foundation on which to perform further clinical sample validation in subsequent research.
The TRAIL pathway is a classically extrinsic signaling pathway that mediates cell apoptosis (23). One study examined tumor cells and discovered that SNAI1 could increase TRAIL-induced apoptosis by enhancing P53 expression (24). In the current study, we discovered that patients with TP53 wild status tended to present with a high expression of SNAI1. This is in line with previous studies in which SNAI1 was frequently overexpressed in BC, inhibiting TRAIL-induced apoptosis and leading to the proliferation of tumor cells. Integrin-related pathways, such as the α9β1 integrin pathway, integrin family cell surface interactions, β1 integrin cell surface interactions, and others, were also significantly enriched by SNAI1 or SNAI2 and their co-expressed genes. This phenomenon may be due to integrins playing key roles in the EMT process (25). For example, integrin α3β1 can bind to laminin and E-cadherin to promote TGFβ-SMAD signaling and induce EMT (26), and increased integrin α5β1 expression can enhance cell adhesion and cell migration (27). Moreover, the mechanism of integrin members promoting EMT is general as they can induce PI3K/AKT pathway activation by increasing SNAI1 expression; receptor tyrosine kinase (RTK)-related pathways, including ErbB receptor signaling and c-Met signaling, work by a similar principle (28). In BC, the loss of hormonal dependency is often accompanied by the appearance of EMT, while the increased expression of SNAI1 is associated with a decrease in estrogen dependency (29). Thus, SNAI1 and its co-expressed genes are enriched in the estrogen receptor (ER) signaling pathway.
Furthermore, compared with the other proteins, SNAI3 and its co-expressed genes were more enriched in immune-related pathways and biological processes. Although SNAI3 shares the same basic structure with the other members in this family and is always well characterized as a transcriptional repressor, these findings indicate that SNAI3 may also play a role in the function of the immune system. In the present study, SNAI3 was highly expressed in skeletal muscle and lymph nodes, which suggests that the deletion of this gene might lead to embryonic lethality and T cell development anomalies. However, researchers previously discovered that SNAI3 knockout in mice had no impact on reproduction, viability, or immune cell derivation (30). Further experiments found that SNAI2 functionally complements the absence of SNAI3, and the analysis of the hematopoietic system in double-knockout mice (SNAI2-/SNAI3-) revealed that these two genes could influence the development of hematopoietic cells (31). Also, the conditional double deletion of SNAI2 and SNAI3 could produce a fatal autoimmunity of severely impairing B cell and T cell generation, and this phenomenon could be reversed by enhancing wild-type Tregs (32). In other words, the Snail family members may act as transcriptional regulators that play necessary roles in maintaining immune tolerance. SNAI3 was revealed to function as a transcriptional regulator in the hematopoietic system; however, SNAI1 was not required for hematopoiesis as SNAI1-deleted mice were healthy and showed no obvious hematopoietic deficiencies (31,33). Meanwhile, the overexpression of SNAI3 in the hematopoietic system led to a decrease of mature lymphocytes and an increase in the development of cells with myeloid lineage. Taken together, these results suggest that SNAI3 mediates the immune process.
Conclusions
In conclusion, the upregulation of the SNAI1 was associated with poor prognosis in BC patients, whereas increased SNAI3 expression was associated with better survival. Based on these findings, further exploration into the exact function of SNAI1 and SNAI3 in BC are warranted.
Acknowledgments
Funding: This research was supported by a grant from the National Nature Science Foundation of China (81802494).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/atm-20-681
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-681
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-681
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-681). The authors have no conflicts of interest to declare.
Ethicals Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Forouzanfar MH, Foreman KJ, Delossantos AM, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet 2011;378:1461-84. [Crossref] [PubMed]
- Fan L, Zheng Y, Yu KD, et al. Breast cancer in a transitional society over 18 years: trends and present status in Shanghai, China. Breast Cancer Res Treat 2009;117:409-16. [Crossref] [PubMed]
- Silber JH, Rosenbaum PR, Clark AS, et al. Characteristics associated with differences in survival among black and white women with breast cancer. JAMA 2013;310:389-97. [Crossref] [PubMed]
- Li J, Zhang BN, Fan JH, et al. A nation-wide multicenter 10-year (1999-2008) retrospective clinical epidemiological study of female breast cancer in China. BMC Cancer 2011;11:364. [Crossref] [PubMed]
- Smith EC, Ziogas A, Anton-Culver H. Delay in surgical treatment and survival after breast cancer diagnosis in young women by race/ethnicity. JAMA Surg 2013;148:516-23. [Crossref] [PubMed]
- de Herreros AG, Peiró S, Nassour M, et al. Snail family regulation and epithelial mesenchymal transitions in breast cancer progression. J Mammary Gland Biol Neoplasia 2010;15:135-47. [Crossref] [PubMed]
- Cano A, Pérez-Moreno MA, Rodrigo I, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2000;2:76-83. [Crossref] [PubMed]
- Bolós V, Peinado H, Pérez-Moreno MA, et al. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci 2003;116:499-511. [Crossref] [PubMed]
- Larue L, Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3' kinase/AKT pathways. Oncogene 2005;24:7443-54. [Crossref] [PubMed]
- Shin NR, Jeong EH, Choi CI, et al. Overexpression of Snail is associated with lymph node metastasis and poor prognosis in patients with gastric cancer. BMC Cancer 2012;12:521. [Crossref] [PubMed]
- Shih JY, Tsai MF, Chang TH, et al. Transcription repressor slug promotes carcinoma invasion and predicts outcome of patients with lung adenocarcinoma. Clin Cancer Res 2005;11:8070-8. [Crossref] [PubMed]
- Katoh M, Katoh M. Identification and characterization of human SNAIL3 (SNAI3) gene in silico. Int J Mol Med 2003;11:383-8. [PubMed]
- Zhuge X, Kataoka H, Tanaka M, et al. Expression of the novel Snai-related zinc-finger transcription factor gene Smuc during mouse development. Int J Mol Med 2005;15:945-8. [PubMed]
- Zhang J, Yan Y, Cui X, et al. CCL2 expression correlates with Snail expression and affects the prognosis of patients with gastric cancer. Pathol Res Pract 2017;213:217-21. [Crossref] [PubMed]
- Chang HY, Tseng YK, Chen YC, et al. High snail expression predicts a poor prognosis in breast invasive ductal carcinoma patients with HER2/EGFR-positive subtypes. Surg Oncol 2018;27:314-20. [Crossref] [PubMed]
- Liu WS, Chan SH, Chang HT, et al. Isocitrate dehydrogenase 1-snail axis dysfunction significantly correlates with breast cancer prognosis and regulates cell invasion ability. Breast Cancer Res 2018;20:25. [Crossref] [PubMed]
- Wang C, Liu X, Huang H, et al. Deregulation of Snai2 is associated with metastasis and poor prognosis in tongue squamous cell carcinoma. Int J Cancer 2012;130:2249-58. [Crossref] [PubMed]
- Pulkka OP, Nilsson B, Sarlomo-Rikala M, et al. SLUG transcription factor: a pro-survival and prognostic factor in gastrointestinal stromal tumour. Br J Cancer 2017;116:1195-202. [Crossref] [PubMed]
- Mendoza-Villanueva D, Balamurugan K, Ali HR, et al. The C/EBPdelta protein is stabilized by estrogen receptor alpha activity, inhibits SNAI2 expression and associates with good prognosis in breast cancer. Oncogene 2016;35:6166-76. [Crossref] [PubMed]
- Pathan M, Keerthikumar S, Ang CS, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015;15:2597-601. [Crossref] [PubMed]
- Chen C, Aihemaiti M, Zhang X, et al. FOXD4 induces tumor progression in colorectal cancer by regulation of the SNAI3/CDH1 axis. Cancer Biol Ther 2018;19:1065-71. [Crossref] [PubMed]
- Madden SF, Clarke C, Stordal B, et al. OvMark: a user-friendly system for the identification of prognostic biomarkers in publically available ovarian cancer gene expression datasets. Mol Cancer 2014;13:241. [Crossref] [PubMed]
- Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer 2008;8:782-98. [Crossref] [PubMed]
- Wan Z, Pan H, Liu S, et al. Downregulation of SNAIL sensitizes hepatocellular carcinoma cells to TRAIL-induced apoptosis by regulating the NF-kappaB pathway. Oncol Rep 2015;33:1560-6. [Crossref] [PubMed]
- Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev 2009;28:15-33. [Crossref] [PubMed]
- Kim Y, Kugler MC, Wei Y, et al. Integrin alpha3beta1-dependent beta-catenin phosphorylation links epithelial Smad signaling to cell contacts. J Cell Biol 2009;184:309-22. [Crossref] [PubMed]
- Mise N, Savai R, Yu H, et al. Zyxin is a transforming growth factor-β (TGF-β)/Smad3 target gene that regulates lung cancer cell motility via integrin α5β1. J Biol Chem 2012;287:31393-405. [Crossref] [PubMed]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014;15:178-96. [Crossref] [PubMed]
- Scherbakov AM, Andreeva OE, Shatskaya VA, et al. The relationships between snail1 and estrogen receptor signaling in breast cancer cells. J Cell Biochem 2012;113:2147-55. [Crossref] [PubMed]
- Pioli PD, Dahlem TJ, Weis JJ, et al. Deletion of Snai2 and Snai3 results in impaired physical development compounded by lymphocyte deficiency. PLoS One 2013;8:e69216. [Crossref] [PubMed]
- Pioli PD, Weis JH. Snail transcription factors in hematopoietic cell development: a model of functional redundancy. Exp Hematol 2014;42:425-30. [Crossref] [PubMed]
- Pioli PD, Chen X, Weis JJ, et al. Fatal autoimmunity results from the conditional deletion of Snai2 and Snai3. Cell Immunol 2015;295:1-18. [Crossref] [PubMed]
- Hale JS, Dahlem TJ, Margraf RL, et al. Transcriptional control of Pactolus: evidence of a negative control region and comparison with its evolutionary paralogue, CD18 (beta2 integrin). J Leukoc Biol 2006;80:383-98. [Crossref] [PubMed]


