
DNA methylation status of imprinted H19 and KvDMR1 genes in human placentas after conception using assisted reproductive technology
Introduction
Assisted reproductive technologies (ARTs), including in vitro fertilization and embryo transfer (IVF-ET) and intracytoplasmic sperm injection (ICSI), have become effective treatments for infertility. Fertility in China has long been constrained by the One-Child policy, enacted in 1978 (1). Worldwide, over 5 million babies have been born via ART since the first assisted conception in 1978 (2). Most babies born using these reproductive technologies are perceivably healthy.
Both IVF-ET and ICSI involve the controlled ovarian hyperstimulation of women, to obtain enough numbers of mature oocytes, followed by zygotic activation, with the first cell divisions occurring in vitro. The artificial control of ovarian hyperstimulation and the culture media used to cultivate germ cells and zygotes may influence the epigenetic reprogramming of ART zygotes. Changes that affect the stability of genomic imprints at differentially methylated regions (DMRs), which control the allelic expression of imprinted genes, are of particular concern (3,4).
ARTs are generally considered to be relatively safe; however, ARTs have been suggested to disturb epigenetic (re)programming during gamete and embryo development, which could have potentially adverse consequences on the epigenetic composition of the embryo (5-7). Recent studies have shown that ART may increase the risk of congenital disabilities, genetic imprinting disorders, and some childhood tumors (8-10).
DNA methylation and DMRs has been the most studied epigenetic modification and is associated to gene silencing when occurring in the promoter region of genes. Imprinted DMRs can influence expression by acting as methylation-sensitive insulators, such as the H19 DMR found on chromosome 11p15.5, the imprinting DMRs can influence gene expression through non-coding RNA (ncRNA) promoters, such as the KvDMR1, also on 11p15.5 chromosome (Vasconcelos, 2019 #65). So we studied the DNA methylation status of imprinted H19 and KvDMR1 genes in human placentas after conception using assisted reproductive technology.
The effects of IVF on DNA methylation were first described in mice (11). Subsequent experiments in mouse embryos demonstrated that imprinted domains, such as the Beckwith-Wiedemann syndrome (BWS) and the Prader-Willi/Angelman syndrome regions, might acquire imprinting errors during IVF (12-14). Smaller epidemiological studies in humans at the beginning of this decade strengthened this view, suggesting a more than three-fold increase in the incidence of BWS among children conceived through ARTs (15,16). These findings were supported by a British survey that observed a slight increase in the frequency of BWS, but not Prader-Willi syndrome (PWS) in ART-conceived children (17). The hypomethylation of KvDMR1, a maternally methylated CpG island found in the promoter of the paternally expressed antisense RNA, termed KCNQ1 overlapping transcript1, represents the most frequent alteration observed in BWS patients.
However, epimutations in ART-conceived children do not appear to be restricted to this locus and may occur at other DMRs, such as those found in the mesoderm-specific transcript (MEST) and insulin-like growth factor 2 receptor (IGF2R) (18). Lim and colleagues observed the increased incidence of DMR hypomethylation in BWS children conceived by ART compared with BWS children who were spontaneously conceived (19). However, extensive epidemiological studies in Denmark, Sweden, and the UK did not observe increased frequencies in the incidence of imprinting disorders among children conceived by ART. They suggested the potential influence of gamete manipulation and embryo culture on DNA methylation at DMRs (19,20). Also, most of the studies that have been performed, thus far, have been biased towards BWS-affected children (20), which may have distorted the views regarding the ART’s influence on genetic imprints because BWS-affected children carry DNA methylation defects at crucial DMRs, by default. Moreover, BWS is still relatively rare; therefore, existing studies have been based on small numbers of BWS-affected children conceived through ART. To circumvent this limitation, Gomes et al. (21) recently conducted a study examining clinically healthy children and found a higher frequency of KvDMR1 hypomethylation among children conceived by ART compared with spontaneously conceived children.
To clearly understand the effects of ART on methylation, we must perform the comprehensive characterization of cells during distinct reprogramming stages, including gene expression profiling and the examination of DNA modifications (22). The DNA methylation statuses of H19 and KvDMR1 were measured in both ART-conceived and naturally conceived newborns to evaluate the safety of ARTs.
Methods
Study design
From June 2018 to December 2018, the placental tissues from 6 full-term deliveries resulting from natural pregnancies and the placental tissues from 6 full-term deliveries resulting from fresh ET pregnancies were collected. The inclusion criteria for all subjects were as follows: single-term fetus; mothers without gestational hypertension, gestational diabetes, hyperthyroidism or hypothyroidism; and no apparent congenital system defects related to embryonic development or neonatal malformations. All procedures performed in studies involving human participants were following the ethical standards of the institutional research committee and with the Declaration of Helsinki (as revised in 2013) and its later amendments or comparable ethical standards. This study was approved by Medical and Life Science Ethics Committee of Tongji University. All patients signed informed consent.
Maternal clinical information was recorded. The fetal body mass was measured at once after delivery, the placenta was weighed, and the placental tissues near the umbilical cord was removed. Placental tissues were immediately rinsed with saline at 4 °C until no blood remained, cut into 1 cm × 1 cm × 1 cm pieces, frozen for 1 h in liquid nitrogen, and stored at −80 °C until use.
Primary reagents and instruments
We extracted genomic DNA (gDNA) and RNA from two sets of specimens. gDNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Fremont, CA, USA). The purified DNA was then quantified and evaluated using a NanoDrop ND-1000. RNA extraction was performed using RNAiso Reagent. The transcription kit and the fluorescent polymerase chain reaction (PCR) kit were all bought from Takara, Japan. Bio-Rad CFX Connect manufactured the real-time PCR instrument. The whole-genome DNA methylation detection chip used was the Arraystar Human RefSeq Promoter Array.
Detecting the mRNA expression levels of genes of interest
Real-time qualitative PCR (RT-qPCR) was used to analyze the mRNA expression levels of H19 and KvDMR1 in the placental sample. β-actin was used as the internal reference gene.
Extracting RNA from placental tissue
For RNA extraction, 50 mg of placental tissue was used, according to RNA extraction kit instructions. After RNA extraction, the RNA concentration and A260/A280 were determined using an ND2000 spectrophotometer. An A260/A280 ratio between 1.9 and 2.1 showed excellent RNA purity.
cDNA synthesis
cDNA synthesis was performed using the Takara reverse transcription kit from Japan using extracted RNA as the raw material. A total of 1 µL RNA was added to the 20 µL system volume, according to the manufacturer’s instructions.
RT-qPCR
The primer sequences were found using Primer Bank and synthesized by Shanghai Shenggong Bioengineering Co., Ltd. For the target geneH19,the forward primer was 5'-GAGCCGCACCAGATCTTCAG-3', and the reverse primer was 5'-TTGGTGGAACACACTGTGATCA-3'; The PCR procedure is “5 min at 94 °C, followed by 35 cycles of 30 sec at 94 °C, 45 sec at 58 °C (β-actin: 58 °C; H19: 60 °C), 30 sec at 72 °C, 72 °C 5 min, 4 °C hold”. For KvDMR1, we obtained the primer sequence from the assay (23). In the first round of PCR, the forward primer LITBisF (5'-GGGGGTTTTTTAGTATGGTTTTTTTT-3' nucleotide position 67934–67959 in GenBank accession number U90095) labelled with 59-indocarbocyanin was used, together with the reverse primer LITRV (5'-CACTACCCAAACCAAACTACACTAC-3'; 68202–68225). In the second round of PCR, the reverse primer LITRV was combined with the 59-indocarbocyanin-labelled forward primer LITFI (5'-GGTTTTTTTTAT TTTTTTGGGAGGGTTTG-3' 68070–68098). The following PCR programme was used for genomic DNA: a denaturation step of 5 min at 94 °C followed by 20 cycles of 30 sec at 94 °C, 30 s at 68 °C and 30 sec at 72 °C, and a final extension for 5 min at 72 °C. A volume of 3 ml of the first round was used as DNA input for amplification in the second round of PCR with the following programme: 5 min of denaturation at 94 °C followed by 25 cycles of 30 sec at 94 °C, 30 sec at 63 °C and 30 s at 72 °C, and a final extension step for 5 min at 72 °C. The PCR programme for single or a few germ cells has an additional five cycles in the first round and ten cycles in the second round.
The reaction system and procedures were performed according to the manufacturer's instructions using the YOBR Premix ExTaq reagent, purchased from Takara Corporation of Japan. Three replicate wells were used for each sample. After the reaction, the amplification curves and melting curves were recorded to determine the specificity of the sample amplification. According to the PCR amplification curves, the number of cycles (CT value) for each sample was obtained, and the relative expression level of the target gene mRNA was calculated using the 2−∆∆CT method.
Whole-genome DNA methylation microarray
Whole-genome DNA methylation microarray was used to examine three placental samples from full-term deliveries resulting from natural pregnancies and three placental samples from full-term deliveries resulting from IVF pregnancies.
To compare the DNA methylation level among the two groups, Methylation 450 microarray data are normalized. Significantly methylated loci showing the change of DNA methylation in whole genome were shown by setting P≤0.05 as a threshold for screening and drawing Circos plot using OmicCircos R package. DNA methylation in different gene regions may cause the difference in the gene expression level and the gene function. The changes of DNA methylation level in different gene regions [gene body, exoniensis 1 (Exon 1), transcriptional start site 1500 (TSS1500), transcriptional start site 200 (TSS200), untranslated region (UTR)-3, and UTR-5] were compared from a system perspective.
Genomic DNA extraction and fragmentation
gDNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Fremont, CA, USA). The purified DNA was then quantified and evaluated using a NanoDrop ND-1000. Using the Bioruptor sonicator (Diagenode), set to “Low” mode for ten cycles (30 seconds “ON” and 30 seconds “OFF”), ultrasound was used to create fragments of approximately 200–1,000 bp. gDNA and sheared DNA were detected using agarose gel electrophoresis.
Immunoprecipitation
A total of 1 µg ultrasound-fragmented gDNA was immunoprecipitated using a 5-methylcytosine mouse monoclonal antibody (Diagenode). DNA was denatured at 94 °C for 10 min and quickly placed on ice before adding 1 µL primary antibody and 400 µL immunoprecipitation buffer [0.5% bovine serum albumin (BSA) in phosphate-buffered saline (PBS)] and mixing, overnight at 4 °C. Recovery of antibody-bound DNA fragments was performed by adding 200 µL of anti-mouse IgG magnetic beads and mixing at 4 °C for 2 h. After the antibody was hybridized, the sample was washed five times at 4 °C. After washing, the magnetic beads were resuspended at 65 °C for 2 h in TE buffer, holding 0.25% SDS and 0.25 mg/mL proteinase K, and then cooled to room temperature. DNA was recovered using Qiagen MinElute columns (Qiagen).
DNA labeling and chip hybridization
The purified DNA was quantified using a NanoDrop ND-1000. DNA labeling was performed using the NimbleGen Dual-Color DNA Labeling Kit. The experimental procedure was based on the standard NimbleGen Methylated DNA immunoprecipitation (MeDIP)-chip protocol. A total of 1 µg DNA and 1 OD Cy5-9mer primer (IP sample) or Cy3-9mer primer (Input sample) were incubated at 98 °C for 10 min. Then, 100 pmol of dNTP and 100 U of Klenow fragment (New England Biolabs, USA) were added and mixed at 37 °C for 2 h. The reaction was ended by the addition of 0.1 volume of 0.5 M EDTA, and the labeled DNA was recovered by is opropanol or ethanol precipitation. The labeled DNA and chip were hybridized at 42 °C for 16–20 h. The hybridization process was performed in a hybridization cassette using the Nimblegen hybridization buffer/hybridization component A (Hybridisation System - Nimblegen Systems, Inc., Madison, WI, USA). After the hybridization was completed, the chip was washed using a Nimblegen Wash Buffer kit (Nimblegen Systems, Inc., Madison, WI, USA).
Promoter differential methylation cluster analysis
The results of differential methylation analysis were clustered to visualize the DNA methylation enrichment peaks for the probes in each sample. Clustering uses log2 ratio value data for the probes in all regions.
Statistical analyses
The expression values and the global hydroxymethylation data was evaluated by the Mann-Whitney U test and the methylation data was evaluated by the t-test, using Graphpad Prsim 7.0. The results of all tests to assess the significance of the observed differences between groups were considered significant when P<0.05.
To eliminate systematic errors and assess differences in methylation between two groups, we performed median normalization, quantile normalization, and linear smoothing of the chip data, using Bioconductor’s Ringo, Limma, and MEDME packages, respectively. When comparing the differential enrichment zones between the two sets of samples, we first calculated the average log2-ratio value of each probe in each set of samples (e.g., test and control) and then calculated the M' values [M' = Average (log2MeDIPE/InputE) – Average(log2MeDIPC/InputC)]. Then, we reran the Nimble Scan sliding window peak-finding algorithm to find differential enrichment peaks (DEPs).
Results
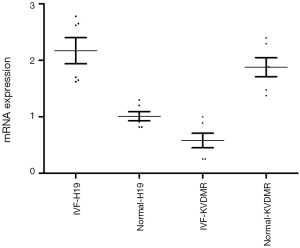
The mRNA expression levels of H19 and KvDMR1 in the IVF group compared with the natural pregnancy group
The expression of level H19 in the IVF group was significantly higher than that in the natural pregnancy group (2.17±0.55 versus 1.01±0.19, P<0.001), whereas the expression level of KvDMR1 was significantly lower in the IVF group than in the natural pregnancy group (0.58±0.32 versus 1.88±0.41, P<0.001) (Figure 1).
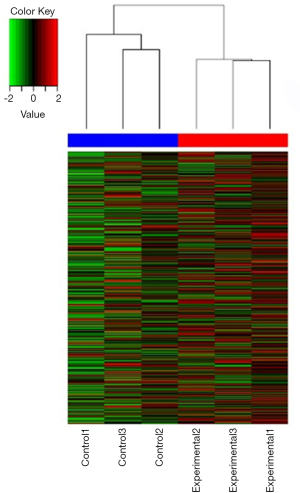
Cluster analysis and differential gene screening
The expressed genes were analyzed by hierarchical clustering to display the gene expression differences comprehensively and intuitively between the two groups. Direct correlations were calculated based on the expression levels of selected genes.
For each experimental group, using more than three biological replicates, we applied a Student’s t-test to find differentially expressed genes. High-CpG-density promoters, low-CpG-density promoters, and intermediate-CpG-density promoters all showed hundreds of DEPs, and several genes with substantial differences were identified. Named hypermethylated genes included GPR85, SPDEF, LAMP2, and ITGB7. Named hypomethylated genes includedTRIM26, VEGFA, CGRRF1, and DAZ-2/3/4 (Figure 2).
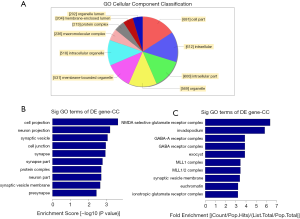
Gene ontology (GO) analysis
GO is used to analyze connections among genes. The results show the top ten valid enrichment terms (Figure 3A). The bar charts show the top ten “rich enrichment” values for the valid enrichment terms (Figure 3B,C). The results of the GO analysis showed that DNA methylation differed significantly depending on the cellular functions and signaling pathways associated with each differentially methylated gene. The results suggested that human ART manipulation may contribute to placental genomic DNA methylation modifications.
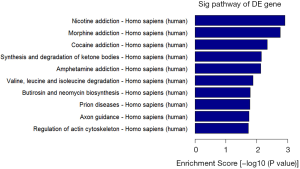
Pathway analysis
The top ten enrichment scores [–log10(P value)] for significant enrichment pathways associated with differentially methylated genes (Figure 4).
Discussion
ART pregnancies have higher risks than naturally conceived pregnancies for several complications and adverse outcomes, including perinatal mortality, preterm birth, intrauterine growth restriction (IUGR), small for gestational age, and congenital abnormalities (24). The reasons for these increases in adverse outcomes during ART pregnancies remain unclear; however, the underlying infertility of these couples, controlled ovarian hyperstimulation, and the actual handling of gametes and embryos could potentially play roles in increasing risks. Recently, the potential effects of ART on the epigenetics of the oocyte/embryo have become the focus of particular concern because ART procedures (controlled ovarian hyperstimulation, in vitro maturation, ICSI, and embryo culture media) are performed during a period associated with epigenetic reprogramming and may be sensitive to epigenetic disturbances (25,26). Alterations in epigenetic mechanisms may contribute to the poor birth outcomes seen for ART pregnancies and could have potential long-term health implications for ART-conceived individuals. Because disorders associated with genomic imprinting have been observed at higher rates for ART-conceived pregnancies compared with naturally conceived pregnancies, the effects of ART with regards to genomic imprinting and DNA methylation have become a focus of concern (27).
This study initially compared the DNA methylation status of the imprinted genes H19 andKvDMR1 in human placentas from full-term natural and ART-assisted pregnancies. Figure 1 shows that the expression level of H19 in the IVF group was significantly higher than that in the natural pregnancy group, whereas the expression level of KvDMR1 was significantly lower in the IVF group compared with the natural pregnancy group. Studies by de Waal et al. (28) have shown that placentas derived from ART have higher frequencies of methylation abnormalities than embryos derived from ART. DNA methylation plays an essential role during mammalian development (29). Katari et al. (30) studied the placenta and cord blood and found that the DNA methylation levels of CpG islands from ART placentas were significantly decreased compared with naturally conceived placentas. In contrast, the DNA methylation levels of CpG islands in ART cord blood were significantly increased compared with naturally conceived cord blood, and these DNA methylation changes resulted in changes in gene expression. Previous studies have shown that the DNA methylation levels of specific essential genes in ART-conceived placentas were significantly downregulated compared with those in naturally conceived placentas.
Placenta samples from 3 naturally conceived pregnancies and 3 IVF pregnancies applied to a whole-genome DNA methylation microarray. We performed GO analysis on the chip results, and cluster analysis of the DEPs for methylated promoters was performed, as shown in Figure 2, to visualize the DNA methylation of the probes in each sample.
Allele-specific DNA methylation was the most crucial imprinting marker localized to DMRs, and the aberrant methylation if imprinted gDNA has been associated with human diseases, including PWS and cancer (31). Numerous studies have confirmed that the transcriptional repression of downstream promoter genes is associated with promoter methylation, and gene promoters with different GC contents in mammals are known to have different methylation profiles. Based on CpG ratios, GC contents, and CpG-rich region lengths, the promoters were divided into the following three categories: High-CpG-density promoters, low-CpG-density promoters, and intermediate-CpG-density promoters. Hundreds of DEPs were found in all three categories, and several genes with substantial DEP differences were selected. The hypermethylated genes included GPR85, SPDEF, LAMP2, and ITGB7. The hypomethylated genes includedTRIM26, VEGFA, CGRRF1, and DAZ-2/3/4.
As shown in Figure 3, we analyzed the associations among these genes using differential methylation gene function (GO) enrichment analysis. The GO analysis of the methylation chip results showed significant differences in DNA methylation, based on cell function. GO analysis is the international standard for the classification of gene functions. The GO enrichment analysis of genes associated with DMR regions can uncover the biological processes involved in the differential methylation. Studies have shown that methylation primarily regulates gene expression through gene promoter regions, and genes with DMRs that coincide with gene promoters (upstream 2-kb regions) were selected for the GO enrichment analysis. GO terms associated with five or more genes, with enrichment multiples greater than or equal to 2, and q-values less than 0.05 were plotted.
In Figure 4, differential methylation signaling pathway Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed to determine the signaling pathway relationships among genes with significant differences in DNA methylation. In vivo, different genes coordinate with each other to exercise their biological functions, and significant enrichment pathways can identify the most essential biochemical metabolic and signal transduction pathways associated with DMR-related genes. KEGG is the central public database used to find pathways. During pathway significant enrichment analysis using the KEGG pathway, hypergeometric tests were used to find pathways that were significantly enriched in DMR-related genes compared with the entire genome background. Earlier results have suggested that ART manipulation can cause changes in placental genomic DNA methylation modifications. Our results showed that ART significantly downregulated the total methylation level of human placental DNA, suggesting that the downregulation of DNA methylation by ART is a common phenomenon throughout the genome. We intended to study the effects of ART on DNA methylation levels and to uncover potential explanations for the observed decreases in total DNA methylation levels following ART. The placenta is an organ that shows maternal contact and exchanges nutrients during pregnancy. Abnormal functional structures not only increase the risks of pregnancy complications, such as low body weight, premature birth, and pre-eclampsia but also pose long-term health risks for mothers and babies (32). Studies in humans and rodents have generally shown that ART causes changes in placental morphologies and molecular expression levels (33).
The results of this study showed that human ART manipulation causes changes in placental genomic DNA methylation modifications. The level of methylation was significantly reduced, and the molecular mechanisms leading to this decrease in methylation were revealed. We explored which specific genes in the placenta cause methylation changes, elucidating the possible biological effects of DNA methylation abnormalities, and ultimately explaining the mechanism through which ART causes abnormal levels of placental DNA methylation. In the future, we will collect more samples to verify the above findings.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (grant number 81720108017). We thank Geno Biotech China Co., Ltd. for supplying technical support during this study.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3364
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3364). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were following the ethical standards of the institutional research committee and with the Declaration of Helsinki (as revised in 2013) and its later amendments or comparable ethical standards. This study was approved by Medical and Life Science Ethics Committee of Tongji University. All patients signed informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li HT, Hellerstein S, Kang C, et al. The number of births in China in 2015: policy meets superstition. Science Bulletin 2018;63:1235-7. [Crossref]
- Cedars MI. In vitro fertilization and risk of autistic disorder and mental retardation. JAMA 2013;310:42-3. [Crossref] [PubMed]
- Fauque P, Jouannet P, Lesaffre C, et al. Assisted reproductive technology affects developmental kinetics, H19 imprinting control region methylation and H19 gene expression in individual mouse embryos. BMC Dev Biol 2007;7:116. [Crossref] [PubMed]
- Sato A, Otsu E, Negishi H, et al. Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum Reprod 2007;22:26-35. [Crossref] [PubMed]
- Huang J, Lu X, Xie Q, et al. Timing of frozen-thawed embryo transfer after controlled ovarian stimulation in a non-elective freeze-all policy. Ann Transl Med 2019;7:752. [Crossref] [PubMed]
- Barragán M, Freour T, Vassena R. Epigenetics, reproduction and assisted reproduction technology. Médecine de la Reproduction 2015;17:152-9.
- Wang Y, Liu Q, Tang F, Yan L, Qiao J. Epigenetic Regulation and Risk Factors During the Development of Human Gametes and Early Embryos. Annu Rev Genomics Hum Genet 2019;20:21-40. [Crossref] [PubMed]
- Liu L, Gao J, He X, et al. Association between assisted reproductive technology and the risk of autism spectrum disorders in the offspring: a meta-analysis. Sci Rep 2017;7:46207. [Crossref] [PubMed]
- Shu L, Xu Q, Meng Q, et al. Clinical outcomes following long GnRHa ovarian stimulation with highly purified human menopausal gonadotropin plus rFSH or rFSH in patients undergoing in vitro fertilization-embryo transfer: a multi-center randomized controlled trial. Ann Transl Med 2019;7:146. [Crossref] [PubMed]
- Eroglu A, Layman LC. Role of ART in imprinting disorders. Semin Reprod Med 2012;30:92-104. [Crossref] [PubMed]
- Yu B, Smith TH, Battle SL, et al. Superovulation alters global DNA methylation in early mouse embryo development. Epigenetics 2019;14:780-90. [Crossref] [PubMed]
- Elhamamsy AR. Role of DNA methylation in imprinting disorders: an updated review. J Assist Reprod Genet 2017;34:549-62. [Crossref] [PubMed]
- Ma Y, Ma Y, Wen L, et al. Changes in DNA methylation and imprinting disorders in E9.5 mouse fetuses and placentas derived from vitrified eight-cell embryos. Mol Reprod Dev 2019;86:404-15. [Crossref] [PubMed]
- Hawes NA, Fidler AE, Tremblay LA, et al. Understanding the role of DNA methylation in successful biological invasions: a review. Biol Invasions 2018;20:2285-300. [Crossref]
- Berntsen S, Söderström-Anttila V, Wennerholm UB, et al. The health of children conceived by ART: 'the chicken or the egg?'. Hum Reprod Update 2019;25:137-58. [Crossref] [PubMed]
- Tenorio J, Romanelli V, Martin-Trujillo A, et al. Clinical and molecular analyses of Beckwith-Wiedemann syndrome: Comparison between spontaneous conception and assisted reproduction techniques. Am J Med Genet A 2016;170:2740-9. [Crossref] [PubMed]
- Hattori H, Hiura H, Kitamura A, et al. Association of four imprinting disorders and ART. Clin Epigenetics 2019;11:21. [Crossref] [PubMed]
- Rossignol S, Steunou V, Chalas C, et al. The epigenetic imprinting defect of patients with Beckwith-Wiedemann syndrome born after assisted reproductive technology is not restricted to the 11p15 region. J Med Genet 2006;43:902-7. [Crossref] [PubMed]
- Lim D, Bowdin SC, Tee L, et al. Clinical and molecular genetic features of Beckwith-Wiedemann syndrome associated with assisted reproductive technologies. Hum Reprod 2009;24:741-7. [Crossref] [PubMed]
- Källén B, Finnström O, Nygren KG, et al. In vitro fertilization (IVF) in Sweden: risk for congenital malformations after different IVF methods. Birth Defects Res A Clin Mol Teratol 2005;73:162-9. [Crossref] [PubMed]
- Gomes MV, Huber J, Ferriani RA, et al. Abnormal methylation at the KvDMR1 imprinting control region in clinically normal children conceived by assisted reproductive technologies. Mol Hum Reprod 2009;15:471-7. [Crossref] [PubMed]
- Gao R, Liu X, Gao S. Progress in understanding epigenetic remodeling during induced pluripotency. Sci Bull (Beijing) 2015;60:1713-21. [Crossref]
- Geuns E, Hilven P, Van Steirteghem A, et al. Methylation analysis of KvDMR1 in human oocytes. J Med Genet 2007;44:144-7. [Crossref] [PubMed]
- Pandey S, Shetty A, Hamilton M, et al. Obstetric and perinatal outcomes in singleton pregnancies resulting from IVF/ICSI: a systematic review and meta-analysis. Hum Reprod Update 2012;18:485-503. [Crossref] [PubMed]
- Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A 2005;102:10604-9. [Crossref] [PubMed]
- Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet 2007;8:253-62. [Crossref] [PubMed]
- Grafodatskaya D, Cytrynbaum C, Weksberg R. The health risks of ART. EMBO Rep 2013;14:129-35. [Crossref] [PubMed]
- de Waal E, Mak W, Calhoun S, et al. In vitro culture increases the frequency of stochastic epigenetic errors at imprinted genes in placental tissues from mouse concepti produced through assisted reproductive technologies. Biol Reprod 2014;90:22. [Crossref] [PubMed]
- Li G, Zhang M, Chen H, et al. Deep pedigree analysis reveals family specific “fingerprint” pattern of DNA methylation for men. Sci Bull (Beijing) 2017;63:7-10. [Crossref]
- Katari S, Turan N, Bibikova M, et al. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet 2009;18:3769-78. [Crossref] [PubMed]
- Liu Y, Li J, Zhou C, et al. Allele-specific genome editing of imprinting genes by preferentially targeting non-methylated loci using Staphylococcus aureus Cas9 (SaCas9). Sci Bull (Beijing) 2019;21:1592-600. [Crossref]
- Burton GJ, Fowden AL, Thornburg KL. Placental Origins of Chronic Disease. Physiol Rev 2016;96:1509-65. [Crossref] [PubMed]
- Vrooman LA, Xin F, Bartolomei MS. Morphologic and molecular changes in the placenta: what we can learn from environmental exposures. Fertil Steril 2016;106:930-40. [Crossref] [PubMed]


