RNF213 gene polymorphism rs9916351 and rs8074015 significantly associated with moyamoya disease in Chinese population
Introduction
Moyamoya disease (MMD) is an idiopathic intracranial arterial disease characterized by progressive bilateral or unilateral occlusion of the supraclinoid internal carotid artery (ICA) and a hazy network of basal collaterals (1,2). As a rare disease, MMD has been described in all races and ethnicities worldwide especially in Asian countries, particularly Japan, where an incidence up to 0.94 per 100,000 has been reported (3). The main pathological changes in the stenotic segment in MMD include fibrocellular thickening of the intima, causing transient ischemic attacks, cerebral infarction, and intracranial hemorrhage; however, the etiology and pathogenesis of MMD remain unclear (4,5).
Epidemiologic studies have shown that approximately 15% of patients had a family history and accumulating evidence suggests genetic factors as important mediators in the development of MMD (6). A previous genome-wide association study (GWAS) by Kamada et al. had identified Ring Finger Protein 213 (RNF213) at chromosome 17q25 as a susceptible gene related to MMD in Japanese populations (7,8). Subsequent studies in Japan, China, and Korea revealed the close relationship between missense mutations in RNF213 and development of MMD (9-11). RNF 213 p.Arg4810Lys (p.R4810K, rs112735431) was most commonly reported to exhibit a strong association with increased susceptibility to MMD (12). Although the mutation rate of RNF 213 p.Arg4810Lys was similar between Japanese, Korean, and Chinese populations (1.4%, 1.3%, and 1.0%, respectively), MMD-related morbidity is markedly different between these populations (nearly 90% in Japanese, 80% in Koreans, and only 23% in Chinese) (13,14). We speculate that other gene polymorphism in RNF213 may also play a vital role in the disease progress. Therefore, to elucidate the underlying genetic pathogenesis, we investigated five RNF213 polymorphism variants in a cohort of Chinese patients with MMD and compared them with a control population. We present the following article in accordance with the STREGA reporting checklist (available at http://dx.doi.org/10.21037/atm-20-1040).
Methods
Patient’s recruitment
A hospital-based case-control design was conducted in the present study. Patients were consecutively recruited from February to May, 2018 among inpatients in Beijing Tiantan Hospital. MMD was diagnosed as per the guidelines established by the Research Committee on Spontaneous Occlusion of the Circle of Willis (Moyamoya Disease) of the Ministry of Health and Welfare of Japan (15). All patients were enrolled based on the presence of clinical ischemic or hemorrhagic symptoms in combination with vascular lesions in magnetic resonance imaging (MRI) or magnetic resonance angiography (MRA). Patients with a history of autoimmune disease, brain neoplasm, meningitis, Down syndrome, head trauma, or other serious illness were excluded. Meantime, controls group were enrolled from healthy examination individuals without a familial history of related diseases. All participants’ age, gender and medical history were recorded. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consents were obtained from all participants in the study, and the study was approved by the Ethics Committee of Beijing Tiantan Hospital.
SNP selection
The common SNPs of RNF213 was selected from the HapMap (http://hapmap.ncbi.nlm.nih.gov/) of the Han Chinese population. Tag SNPs, including rs35692831, rs9916351, rs9913636, rs8074015 and rs112735431, across the gene were subsequently selected using the Haploview software version 4.2. The linkage disequilibrium r2 threshold was set as 0.8 and the minor allele frequency (MAF) was set to >0.05.
Genotyping
Blood samples were collected from elbow veins of participant and stored at −80 °C until use. Genomic DNA was extracted using a commercial kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. DNA quantity was examined by a NanoDrop2000 spectrophotometer (Thermo Fisher, Waltham, MA, USA). Polymerase chain reaction (PCR) primer pairs used to amplify RNF213 polymorphism were showed in Table 1. Genotyping was performed by using time-of-flight mass spectrometry on a MassARRAY iPLEX platform (Sequenom, San Diego, CA, USA) by Bio Miao Biological Technology (Beijing, China). The average genotype call rate for this SNP was >98%.
Full table
Statistical analysis
All statistical analyses were performed by SPSS (version 17.0, SPSS Inc., Chicago, USA) and P-Link. Hardy-Weinberg equilibrium (HWE) was assessed using a χ2 test and a P value <0.05 was considered to indicate significant disequilibrium. Differences in genotypic frequencies between groups were calculated by the Pearson’s χ2 test or Fisher’s exact test, as appropriate. Odds ratio (OR) values with 95% confidence interval (CI) were calculated. Univariate and multivariable logistic regression analysis were used to evaluate the impact of clinical variables on RNF213 polymorphism status. Patients’ age and gender were treated as risk factors to be adjusted in the analysis. Haplotype was analyzed using online web tools Haploview and SHEsis (http://analysis.bio-x.cn/myAnalysis.php). All tests were two-tailed, and P<0.05 indicated statistical significant.
Results
The characteristic of the study
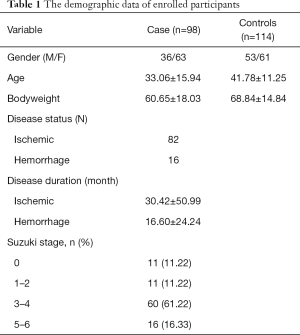
A total of 98 patients (63 female) with mean age 33.06±15.94 and 114 healthy controls (61 female) with mean age 41.78±11.25 were enrolled in this hospital-based case and control study. The average body weight in patient and control group was 60.65±18.03 and 68.84±14.84 kg, respectively. Of them, 82 patients were ischemic type with a duration 30.42±50.99 months, and only 16 patients were hemorrhagic type with a duration 16.60±24.24 months. All participants were Chinese. A total of five tag SNPs were selected for further analysis. The MAF were 0.414, 0.413, 0.289 and 0.322 for rs35692831, rs9916351, rs9913636, rs8074015, and rs112735431, respectively. The demographics characteristics are shown in Table 1.
Genetics association analysis
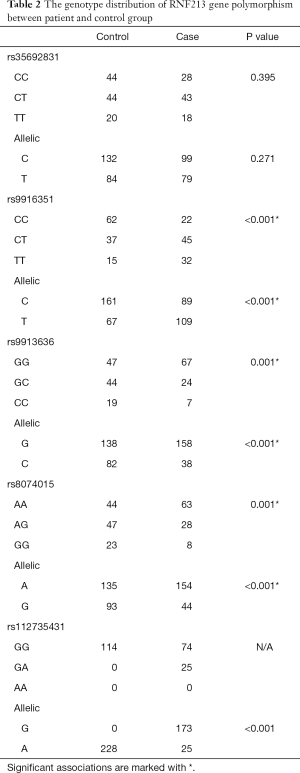
The frequencies of genotype and alleles of the RNF213 gene polymorphisms in patients and healthy controls are shown in Table 2. A significant difference was observed between patients and healthy controls in rs9916351, rs9913636, and rs8074015loci under three genotypes and allelic models (P<0.01). In addition, rs112735431 polymorphism significantly differed only under the allelic genotype (P<0.001). No other significant difference was reported under different genotypes in rs35692831 (P>0.05).
Full table
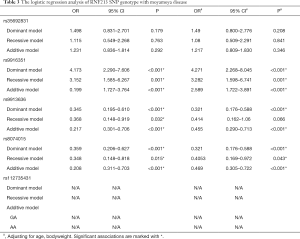
A logistic regression analysis of five SNP genotypes under the dominant, recessive, and additive model between patients and healthy controls was also performed. The results showed significant differences under the dominant and recessive models in rs9916351 (OR =4.173, 95% CI: 2.290–7.606, P<0.001; OR =3.152, 95% CI: 1.585–6.267, P=0.001; respectively) and rs8074015 (OR =0.359, 95% CI: 0.206-0.627, P<0.001; OR =0.348, 95% CI: 0.148–0.81, P=0.015; respectively), even adjusting for age and gender (Table 3). We also observed a significant difference in rs9913636 under the dominant (OR =0.345, 95% CI: 0.195–0.610, P<0.001), and recessive model (OR =0.368, 95% CI: 0.148–0.919, P=0.032). However, no significant difference was observed under the recessive model after adjusting for age and gender. The additive models of rs9916351 (OR =0.199, 95% CI: 1.727–3.764, P<0.001), rs9913636 (OR =0.217, 95% CI: 301–3.764, P<0.001) and rs8074015 (OR =0.208, 95% CI: 0.311–0.703, P<0.001) were also found with significantly difference between MMD patients and healthy control even after adjusting for age and gender (OR =2.589, 95% CI: 1.722–3.891, P<0.001; OR =0.455, 95% CI: 0.290–0.713, P<0.001; OR =0.469, 95% CI: 0.305–0.722, P<0.001).
Full table
Haplotype analysis
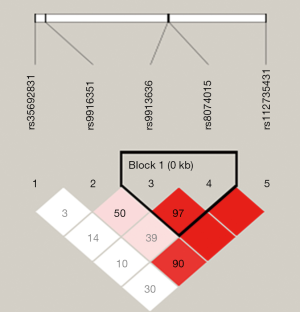
The pairwise linkage disequilibrium D’ values among the five SNPs were calculated for different sample sets. The SNPs with D’ >0.75 in separated sample sets were grouped in the same block, and rs9913636-rs8074015 was identified as one haplotype block (Figure 1). In this block, one haplotype “GACG” showed significant association with MMD (Table 4).
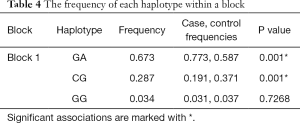
Full table
Discussion
MMD was considered as a chronic cerebrovascular disorder since it was first described by Shimizu and Takeuchi in 1955 in Japan (16). Not until 2011, Kamada et al. revealed a founder missense mutation in RNF213 was tightly associated with MMD onset in Japanese family (7). Since then, several countries particularly in Asia, have reported same results (17). However, as racial differences, MMD incidence are high in Asians, especially in Japanese population, and etiology is still unclear.
The polymorphism of RNF213 was recognized as a susceptible factor to MMD and has been widely studied. Several studies supported that RNF213 p.R4810K variant is common in early-onset ischemic stroke with anterior circulation stenosis (17,18). However, RNF213 p.R4810K variant is not the only susceptibility variant for MMD. To date, at least 24 genetic changes in the RNF213 gene have been found to be associated with MMD (9). In our study, we had observed five SNPS in RNF213 including rs35692831, rs9916351, rs9913636, rs8074015, and rs112735431, all of which were related to MMD morbidity. Our results show that rs9916351 and rs8074015 polymorphisms were closely related to the development of MMD both under dominant and recessive model. To the best of our knowledge, this is the first study to report that rs9913636 in RNF213 is also a susceptibility locus of MMD; nonetheless, further verification of this result in a large case-control study is warranted.
Our results of rs9916351 and rs112735431were in accordance with Duan et al. in a GWAS and Huang et al. in a case control study in Chinese population (13,17). As surgical revascularization is the main treatment regimen for MMD to prevent complications, the relationship between polymorphism and long-term clinical outcome is elusive. Ge et al. reported that the p.R4810Kvariant may not be associated with long-term clinical outcomes in Chinese patients with MMD (19).
Although the polymorphism of RNF213 has been widely detected in patients with MMD, the biochemical function and pathological role of RNF213 have not been completely clarified. RNF213 gene encodes a protein with 5256 amino acid residues that is involved in cellular processes, DNA repair, and signal transduction pathways (20,21). As a RING finger protein with E3 ubiquitin ligase activity and energy-dependent unfoldase activity, RNF213 protein can ubiquitinate specific target proteins and mediate their degradation by proteasomes (22). Single-nucleotide polymorphism of RNF213 may result in disruption of ATP hydrolysis cyclicity, inhibition of angiogenesis, and reduction of antiangiogenic activity of interferon beta-1. The consequences include endothelial cell dysfunction, smooth muscle cell dysfunction, and abnormal hemostasis, all of which could aggravate the proliferation of smooth muscle cells (SMCs) and result in vascular stenosis (9). Liu et al. showed that RNF213 p.R4810K knockout zebrafish presented abnormal sprouting vessels in the head region (23). In addition, Kobayashi et al. revealed that mutation may be correlated with angiogenic capacities and caused people more susceptible to insults of cerebral hypoxia (8). In our study, we had found another two loci polymorphisms (rs9916351and rs8074015) were also significantly corelated with the morbidity of MMD. Thus, in future, more studies focused on the mechanism of these two polymorphisms with the disease are needed.
Although our results are potentially useful to identify MMD patients among the Chinese population, these are preliminary observations and additional studies are deemed imperative. Our study has a few limitations. First, the mechanism of MMD is very complex, and we only observed five SNPs; hence, these SNPS may not completely explain the pathogenesis of this disease. Second, the small sample size of MMD cases may introduce bias. Third, only 16 hemorrhage patients were included in our study, making it difficult to verify the role of polymorphism in hemorrhagic MMD.
Conclusions
In summary, our results reveal the association of RNF213 rs9916351, rs9913636, rs8074015, and rs112735431 polymorphism with MMD in Chinese population. However, these data need to be validated in a larger cohort of patients. Furthermore, functional studies are necessary to investigate whether and how these polymorphisms influence the key pathways involved in the pathogenesis of MMD.
Acknowledgments
The authors appreciate all the participants and their relatives in the study and the members of the survey teams.
Funding: This study was supported by China Postdoctoral Science Foundation (No. 2017M620700), the Natural Science Foundation of China (No. 81803909) and Beijing Municipal Administration of Hospitals (ZYLX201827).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-1040
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-1040
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-1040). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consents were obtained from all participants in the study, and the study was approved by the Ethics Committee of Beijing Tiantan Hospital.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Huang S, Guo ZN, Shi M, et al. Etiology and pathogenesis of Moyamoya Disease: An update on disease prevalence. Int J Stroke 2017;12:246-53. [Crossref] [PubMed]
- Fujimura M, Bang OY, Kim JS. Moyamoya Disease. Front Neurol Neurosci 2016;40:204-20. [Crossref] [PubMed]
- Kuroda S, Houkin K. Moyamoya disease: current concepts and future perspectives. Lancet Neurol 2008;7:1056-66. [Crossref] [PubMed]
- Bersano A, Guey S, Bedini G, et al. Research Progresses in Understanding the Pathophysiology of Moyamoya Disease. Cerebrovasc Dis 2016;41:105-18. [Crossref] [PubMed]
- Shang S, Zhou D, Ya J, et al. Progress in moyamoya disease. Neurosurg Rev 2020;43:371-82. [Crossref] [PubMed]
- Zhang H, Zheng L, Feng L. Epidemiology, diagnosis and treatment of moyamoya disease. Exp Ther Med 2019;17:1977-84. [PubMed]
- Kamada F, Aoki Y, Narisawa A, et al. A genome-wide association study identifies RNF213 as the first Moyamoya disease gene. J Hum Genet 2011;56:34-40. [Crossref] [PubMed]
- Kobayashi H, Matsuda Y, Hitomi T, et al. Biochemical and Functional Characterization of RNF213 (Mysterin) R4810K, a Susceptibility Mutation of Moyamoya Disease, in Angiogenesis In Vitro and In Vivo. J Am Heart Assoc 2015;4:e002146. [Crossref] [PubMed]
- Park YS, An HJ, Kim JO, et al. The Role of RNF213 4810G>A and 4950G>A Variants in Patients with Moyamoya Disease in Korea. Int J Mol Sci 2017;18:2477. [Crossref] [PubMed]
- Ge P, Ye X, Liu X, et al. Association between p.R4810K Variant and Postoperative Collateral Formation in Patients with Moyamoya Disease. Cerebrovasc Dis 2019;48:77-84. [Crossref] [PubMed]
- Sato Y, Kazumata K, Nakatani E, et al. Characteristics of Moyamoya Disease Based on National Registry Data in Japan. Stroke 2019;50:1973-80. [Crossref] [PubMed]
- Fujimura M, Sonobe S, Nishijima Y, et al. Genetics and Biomarkers of Moyamoya Disease: Significance of RNF213 as a Susceptibility Gene. J Stroke 2014;16:65-72. [Crossref] [PubMed]
- Duan L, Wei L, Tian Y, et al. Novel Susceptibility Loci for Moyamoya Disease Revealed by a Genome-Wide Association Study. Stroke 2018;49:11-8. [Crossref] [PubMed]
- Liu W, Hitomi T, Kobayashi H, et al. Distribution of moyamoya disease susceptibility polymorphism p.R4810K in RNF213 in East and Southeast Asian populations. Neurol Med Chir (Tokyo) 2012;52:299-303. [Crossref] [PubMed]
- Research Committee on the P. Treatment of Spontaneous Occlusion of the Circle of W, Health Labour Sciences Research Grant for Research on Measures for Infractable D. Guidelines for diagnosis and treatment of moyamoya disease (spontaneous occlusion of the circle of Willis). Neurol Med Chir (Tokyo) 2012;52:245-66. [Crossref] [PubMed]
- Hu J, Luo J, Chen Q. The Susceptibility Pathogenesis of Moyamoya Disease. World Neurosurg 2017;101:731-41. [Crossref] [PubMed]
- Huang Y, Cheng D, Zhang J, et al. Association between the rs112735431 polymorphism of the RNF213 gene and moyamoya disease: A case-control study and meta-analysis. J Clin Neurosci 2016;32:14-8. [Crossref] [PubMed]
- Kamimura T, Okazaki S, Morimoto T, et al. Prevalence of RNF213 p.R4810K Variant in Early-Onset Stroke With Intracranial Arterial Stenosis. Stroke 2019;50:1561-3. [Crossref] [PubMed]
- Ge P, Ye X, Liu X, et al. Association Between p.R4810K Variant and Long-Term Clinical Outcome in Patients With Moyamoya Disease. Front Neurol 2019;10:662. [Crossref] [PubMed]
- Ma YG, Zhang Q, Yu LB, et al. Role of Ring Finger Protein 213 in Moyamoya Disease. Chin Med J (Engl) 2016;129:2497-501. [Crossref] [PubMed]
- Miyawaki S, Imai H, Takayanagi S, et al. Identification of a genetic variant common to moyamoya disease and intracranial major artery stenosis/occlusion. Stroke 2012;43:3371-4. [Crossref] [PubMed]
- Park YS. Single Nucleotide Polymorphism in Patients with Moyamoya Disease. J Korean Neurosurg Soc 2015;57:422-7. [Crossref] [PubMed]
- Liu W, Morito D, Takashima S, et al. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS One 2011;6:e22542. [Crossref] [PubMed]