Current challenges for detection of circulating tumor cells and cell-free circulating nucleic acids, and their characterization in non-small cell lung carcinoma patients. What is the best blood substrate for personalized medicine?
Introduction
In recent years, lung cancer has been the leading cause of cancer-related mortality among men and women worldwide. It is estimated that there will be 160,000 deaths due to lung cancer in 2014 for the USA alone (1). Moreover lung cancer has a 5-year overall survival rate of 16% for all stages and survival has only improved minimally in the last decades (1). In recent years, a better understanding of the molecular abnormalities present in lung cancer that define disease subsets has been reached. In particular, the discovery of oncogenic drivers has led to the design of therapies harboring specific gene alterations that cause aberrant signaling and proliferation. Currently, most patients with lung cancer routinely undergo a tissue (cellular) biopsy for molecular profiling of their tumors. However, the main challenge of targeted therapies is the relatively small proportion of patients that can benefit from these treatments and the almost inevitable occurrence of resistance (2,3). Some of the resistance can be detected from “solid biopsies”. However, this cannot always be performed because of the invasive characteristic and may fail to reflect current tumor dynamics and drug sensitivity, which may change during therapy. Therefore, the importance of developing a noninvasive biomarker with the ability to monitor in real-time the dynamics of the lung cancer should be emphasized. This “liquid biopsy” should provide an ideal therapeutic strategy for an individual cancer patient, which would facilitate the development of “tailor-made” cancer management programs (4,5).
The liquid biopsy is a noninvasive approach that detects diagnostic, prognostic and theranostic biomarkers in non-small cell lung carcinoma (NSCLC) patients (6). This attractive approach is currently being developed by many investigators, particularly with the aim of obtaining a complementary tool to the solid biopsy for detection of genomic alterations in metastatic cancer patients, which allows delivery of targeted therapy. Moreover, a liquid biopsy can easily be repeated during follow-up of cancer patients to control treatment efficiency and/or the detection of genomic alterations occurring as a result of resistance to targeted therapy. Finally, the liquid biopsy is a tool that allows rapid access to biomarker assessment in vulnerable lung cancer patients for whom solid biopsies are inaccessible or extremely tricky to perform and to repeat. In theory, this approach can promote a change in therapy, even before detection of tumor progression or relapse.
The term “liquid biopsy” qualifies different potential approaches for detecting of blood-carrying biomarkers in lung cancer patients. Some of these biomarkers have the potential to be introduced in the near future into daily clinical practice. However, its success is dependent on robust validation in sufficiently large independent prospectively designed studies. Stricto sensus, the term “liquid biopsy” should be restricted to a blood test that is associated with cytopathological assessment of circulating tumor cells (CTCs) by analogy to the definition of a “tissue” biopsy (National Cancer Institute, NIH, Bethesda, MD). However, under the label “liquid biopsy” different blood-based biomarkers can be detected such as CTCs but also circulating tumor DNA (ctDNA), circulating RNA or microRNAs (7). In addition to obtaining useful data for the care of lung cancer patients, the information obtained from the liquid biopsies should allow an increase into the knowledge of the pathophysiology of lung cancer and into the process of metastatic dissemination.
The aim of this review is to describe the current contribution of detection of CTCs and ctDNA in lung cancer patients and to compare the advantages and disadvantages of these approaches.
Detection of CTCs in lung cancer: current interest and main limitations
For several decades, a lot of translational and clinical lung cancer research programs in the field of CTC have been conducted. Recently, the development of personalized medicine and of the concept of targeted therapy, in particular in lung cancer, has been associated with an increased interest in CTC detection and characterization. In this context, different methods for cellular enrichment and characterization of the identity of the subpopulations of interest have been developed. These methods aim to demonstrate, by direct and indirect approaches, the morphological and molecular characteristics of CTCs in lung cancer patients. Assessment of these methods integrates evaluation of their sensitivity and specificity, their costs and the feasibility of set up and developed not only in research laboratories, but also in hospital biology laboratories for rapid transfer to the clinic.
Currently developed methods used for lung cancer patients
The current technologies are based on exploiting the physical and biological properties of CTCs (8-11). A number of innovative technologies to improve the detection of CTCs have recently been developed, including CTC microchips, filtration devices, quantitative reverse-transcription PCR assays, and automated microscopy systems (12-14). Studies into the molecular characterization have indicated, however, that CTCs are very heterogeneous, a finding that underscores the need for multiplex approaches to capture all the relevant CTC subsets (15). The current challenge is to increase the yield and detection of CTCs that have undergone epithelial-mesenchymal transition (16). However, increasing the sensitivity of the analytical assay may lead to a decrease in the analytical specificity (e.g., through the detection of circulating normal epithelial cells). Despite the effort made by the scientific community in the CTC field, which aims at improving the management of lung cancer, the analytical specificity and clinical utility of these methods must still be demonstrated in large prospective multicenter studies to obtain the high level of evidence of performance required for introduction into the routine clinical practice.
Main interests in pathophysiology, diagnosis, prognosis and theranosis
The knowledge into the pathophysiology of the natural history of lung cancer has improved a lot through different studies concerning CTCs (13). In particular, recent work performed by the team of Massagué in New York has established that CTCs can give rise not only to metastases, but can also colonize the primary tumor site and participate in primary tumor progression, through a phenomenon called tumor self-seeding (17). Other studies showed that CTCs can circulate in the bloodstream of lung cancer patients as single cells or as aggregates [called circulating tumor microemboli (CTMs)]. In this regard, the phenotype of single or aggregated CTCs can be different and may present different levels of potential aggressiveness.
The diagnosis of metastases of unknown primary origin could be determined by the genomic analysis of CTCs. However, the different methods required for such a diagnosis are difficult to set up in routine clinics, and are currently probably too expensive.
Several studies have correlated the presence and the number of CTCs with worse prognosis in lung cancer patients. In early stage lung cancer, the pre-operative detection of CTCs is associated with worse disease free and overall survival (18,19). The detection of CTCs in late stage lung cancer patients and of a high persistent rate of CTCs after chemotherapy in these patients correlated with worse prognosis (15,20).
The detection of some genomic alterations present in CTCs of cancer patients can be associated with targeted therapy (21). Thus, patients with EGFR mutations or EML4-ALK rearrangements detected in CTC can potentially benefit from treatment with tyrosine kinase inhibitors or crizotinib, respectively (22,23).
The main limitations
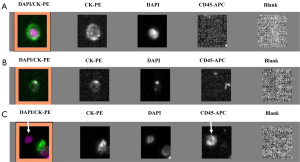
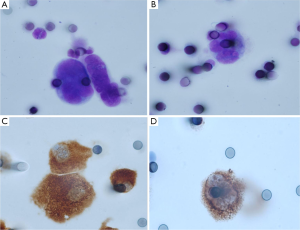
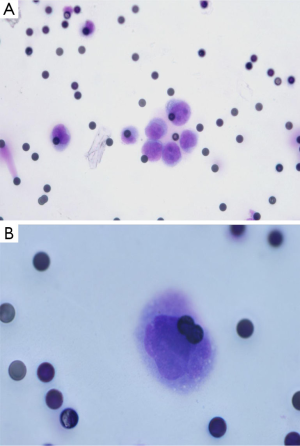
Despite the numerous scientific publications related to CTC detection in lung cancer patients, the physician does not use this biomarker in routine clinical practice. This can be explained by the large number of methods available for CTC detection and by the difficulty of the physician and the biologist to select the optimal method for use (24). In this context, it is noteworthy that the only FDA approved method for CTC detection, the CellSearch method (Janssen Diagnostics Company, USA), has been approved for CTC detection in metastatic breast, prostate and colon cancer patients, but not in metastatic lung cancer patients. This has added some confusion to the use of this indirect technology for lung cancer patient CTC detection. Moreover, since CTCs that have undergone epithelial-mesenchymal transition cannot express epithelial biomarkers, the CellSearch system can certainly miss the detection of a subpopulation of CTCs of interest in lung cancer patients (Figure 1). Direct technologies for CTC detection in lung cancer, such as the ISET approach (developed by Rarecells Company, France), are certainly strongly attractive, but the results obtained by different teams probably need to be validated in independent and large multicenter studies. ISET allows cytological characterization of the CTCs isolated from lung cancer patients, according to the classical morphological criteria used by the cytopathologists for distinguishing between benign and malignant epithelial cells (25,26) (Figure 2). Moreover this approach allows an immunocytochemical assessment of CTCs expressing epithelial and/or mesenchymal biomarkers (Figure 2). Other direct technology, such as those developed by ScreenCell company in France, allows morphological assessment of CTCs in lung cancer patients too (Figure 3). Many other methods are currently being developed for CTC characterization, such as a method allowing functional evaluation the CTCs and characterization of a subpopulation of malignant cells (27). A couple of approaches are sophisticated and give complex data. Currently these new methods seem to be difficult for translation into the clinical routine practice. These approaches lack a multicenter assessment program, and thus it is difficult to evaluate their reproducibility, their sensitivity and their specificity. The development of new technologies and methods for CTC detection is in general not associated with clinical validation in different cohorts of lung cancer patients, making the findings difficult to interpret for the pneumologists.
Detection of ctDNA in lung cancer patients: Interest and current limitations
Two main mechanisms for the release of free-circulating nucleic acids (fcNAs), called “passive” and “active”, have been advocated to date (28). The passive mechanism involves the release of nucleic acids from apoptotic and necrotic cells into the bloodstream. Macrophages and phagocytes play an important role in phagocytosis of necrotic and apoptotic cells and can release digested nucleic acids into the microenvironment. In contrast, it is reported that fragments of cellular nucleic acid can be actively released. Although the active secretion into the circulation remains enigmatic, one potential explanation hypothesizes that cancer cells release nucleic acids to transform the targeted recipient cells at distant locations. In addition to these two mechanisms, cell-free nucleic acids (cfNA) may be released by CTCs; however, there appears to be a huge gap between the amount of cfNAs and the rarity of CTCs in the bloodstream. Thus, this hypothesis remains controversial.
Lung cancer patients demonstrated an increased fcDNA levels in the plasma or serum compared to healthy individuals (29,30). In this regard the level of fcDNA can have a potential interest for the diagnosis and the prognosis of lung cancer and for monitoring of disease during follow-up (31-33). The level of fcDNA seems to be associated with the tumor stage, size and metastases. In this context, a high fcDNA level has been proven to be a biomarker of poor outcome in lung cancer patients (33,34). However, one limitation of these different approaches is that fcDNA is present at a high level in blood patients having benign diseases such as hepatic disorders, diabetes, cardiovascular diseases, non-neoplastic lung diseases or infections (35-37).
The detection of ctDNA in the plasma or the sera of lung cancer patients is also an attractive approach which allows the identification of genomic alterations that can be targeted by different molecules. Many works have shown correlation between genomic alterations (in particular, different EGFR mutations) in lung cancer tissues DNA and its corresponding ctDNA. The high number of recent studies and general reviews concerning this subject highlight the enthusiasm of research scientists and of physicians in exploiting the liquid biopsy of lung cancer patients for targeted therapy development (38-41). Moreover, ctDNA can complement current invasive biopsy approaches to identify mutations associated with acquired drug resistance in advanced lung cancers (42,43) (Figure 4). Thus, the development of personalized medicine through liquid biopsy assessment for cfNAs seems to be competitive in comparison with detection of CTCs. However, one limitation when working with cfDNA samples is the impossibility of doing in situ and morphological analyses using fluorescent in situ hybridization (FISH) and immunocytochemistry (ICC), in particular for the assessment of the ALK, ROS1 and c-MET status.
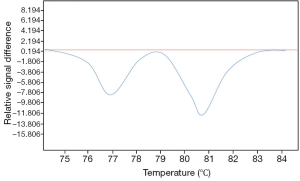
It is noteworthy to consider that despite numerous translational research programs and some clinical trials, the detection of ctDNA in lung cancer patients is currently not used by physicians in daily practice. Thus, the different methods of detection and of analysis have not yet lead to robust commercialized biological tests. Additionally, accreditation for such tests in a biological laboratory seems premature. One of the major constraints of this type of detection is certainly the control of the pre-analytical phase, e.g., the control of the different parameters existing from blood sampling to the analysis of the ctDNA (44-48). Thus, the many steps between these two events can strongly influence the quality and the accuracy of the data. Moreover, analytical phase can be also critical to be evaluated. Different methods can be used for fcDNA quantification. Real time PCR-based methods can be used, but it is pivotal that the target sequences be as short as possible, allowing amplification of shorter fragments. Many methodologies for genomic alteration analysis are available, such as digital PCR, mutant-enriched PCR and peptide nucleic acid locked nucleic acid PCR which are particularly appropriate for fcDNA mutation analysis (49,50). Moreover, next generation sequencing technologies have been recently developed showing that it is possible to detect many cancer-associated mutations fcDNA isolated from plasma of lung cancer patients (51).
CTCs versus ctDNA detection in lung cancer patients: exclusive or complementary approaches?
Since many studies are currently performed either for detection of CTCs or ctDNA in lung cancer patients one can question the efficiency of these two approaches in particular in the area of personalized medicine. Additionally, is it legitimate or not to try to detect in parallel for the same patient, both CTCs and ctDNA?
What information can be obtained exclusively from CTC detection and analysis in lung cancer patients?
Direct technologies for CTC detection can gain information concerning the morphology of the CTC. In this regard, as for cytopathology performed on smears or on fine needle aspiration, CTCs can be identified efficiently as malignant or benign cells according to the established cytological criteria. Previous studies have distinguished different categories of CTCs in lung cancer patients according to these criteria. Thus, the so called “circulating non hematological cells (CNHC)” can show features of malignant or benign cells. Moreover, a couple of CNHC cannot be classified as benign or malignant cells and were called CNHC with uncertain features of malignancy (26). These morphological analyses are pivotal since it is possible to correlate these rare isolated cells with malignant criteria in the blood lung cancer patients. Moreover, ICC and FISH can be performed on these CNHC to obtain better characterization of their phenotype and to detect some genomic alterations accessible to targeted therapy. ICC showed substantial heterogeneity of CTCs in lung cancer patients. As an example, CTCs can express cytokeratin, while other cells express both cytokeratin and vimentin, and finally some CTCs with malignant features can express vimentin only. The direct methods also provide interesting data to better understand the pathophysiology of progression and dissemination of lung cancer. It has been demonstrated that these CTCs can circulate as isolated cells or as aggregates called CTMs. It seems that the presence of CTMs is a negative prognostic factor for lung cancer patients. Interestingly, it is possible to assess the ALK status (by ICC or FISH) of CTCs isolated and characterized by ISET (17,52). Similar approaches could potentially evaluate the EGFR, BRAF, ROS-1 or c-MET status of CTCs isolated by ISET in lung cancer patients since specific antibodies and probes targeting these molecules are now commercially available (53-57). One current drawback is the lack of a biomarker to confirm the malignant nature of CNHCs isolated by the direct methods. Moreover, it is impossible to determine the aggressiveness and the invasiveness of these cells. One of the major pitfalls of many indirect methods for detection of CTCs is the fact that they are based on the postulate that CTCs express an epithelial phenotype. For example the CellSearch technology, which uses anti-Epcam and anti-cytokeratins antibodies for CTC isolation, cannot detect CTCs showing epithelial-to-mesenchymal transition, a frequent process in lung cancer progression and dissemination. Thus, most of the indirect methods can give false negative results.
One of the main difficulties, when working on CTCs in the field of lung cancer, is to analyze the somatic mutations, which can be present in these cells. This is due to the fact that the quantity and the quality of the extracted DNA from the cells are low. In this regard the pre-analytical phase of DNA accessibility can have a strong impact on the quality of the DNA.
What information can be obtained exclusively from detection of free ctDNA and analysis in lung cancer patients?
It is certain that the detection of different somatic mutations (in particular in EGFR) from ctDNA is an exciting area for development in lung cancer patients (58-61). Detection can be performed by different molecular methods of different sensitivity, including targeted methods or more sophisticated approaches such as next generation sequencing (51). These different approaches allow detection of genomic alterations accessible to targeted therapies. It is possible that, the detection and the level of some mutations can represent a negative prognostic factor, as demonstrated in colon cancer patients (62). However a number of difficulties can be highlighted when working on programs concerning ctDNA: (I) it is critical to control well all the parameters of the pre-analytical phase and currently it seems hard, from a clinical setting point of view, to be sure of obtaining robust control for daily practice; (II) the quantity of ctDNA can be too low to develop high throughput analyses; (III) a low amount of DNA can be associated with some artefactual mutations (leading to false positive results); (IV) according to the sensitivity of the method, the presence of a low level of mutated DNA in the whole DNA extract can lead to some false negative results (46,63-65). Among these potential pitfalls, it seems that studying ctDNA in lung cancer patients does not give useful information concerning the degree of tumor aggressiveness.
Conclusions
Although the concept of “liquid biopsy” possesses great potential in detection and monitoring of diseases, as previously described in detail, several hurdles still exist such as the lack of consensus in technical approaches of choice, which involves various aspects of the methodologies, such as the preferable sample type, storage conditions, candidate molecules and suitable detection techniques. Moreover, technical errors may introduce contaminating cells or molecules into samples, which could result in incorrect interpretation and statistical errors. Therefore, the standardization of all experimental steps of techniques should be emphasized. Despite the numerous approaches and techniques that have been advocated to achieve the ultimate goal, that is, the development of a useful, sensitive and real-time monitoring system using blood, few proposals have been translated into the clinical practice. Large-scale studies and further understanding of their biology and significance could resolve these problems and enhance their utility as biomarkers. Consequently, the development of novel biomarkers based on CTCs and cfNAs could provide a lot of benefit to cancer patients, including the improvement in the clinical outcome in the near future.
It is obvious that the concept of a liquid biopsy in the lung cancer field is highly attractive and could allow dramatic optimization of the clinical care of these patients. Moreover, many recent innovative programs aim to use sophisticated technology for circulating single tumor cells or free circulating DNA characterization (66-70). These exciting approaches open new avenues in thoracic oncology. However, behind the technical prowess, one can raise the question of the rapid transfer of technology into the clinical routine practice. As both CTC and ctDNA methods evolve, they will likely have similar but also distinct clinical applications, reflecting their relative biologic and technologic strengths and limits (Table 1).
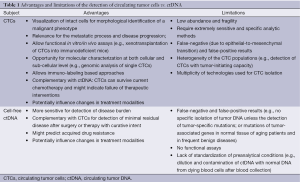
Full table
Of note, the liquid biopsy can allow detection, besides CTCs and ctDNA, of other biomarkers of interest to lung cancer patients, such as circulating microRNAs, circulating RNA or circulating endothelial cells (71,72). However, the results concerning these biomarkers need to be validated in independent cohorts of patients and the “clinical relevance” in the daily practice seems difficult to determine. The rapid changes in the ability to genotype lung cancers and measure their evolving functional properties through noninvasive blood monitoring strongly links lung cancer therapeutics and diagnostics. However, we can ask the following provocative question: when and how the “liquid biopsy” will be used routinely for lung cancer patient care? Currently a real gap exists between the genuine attraction of obtaining a liquid biopsy in media and the increase in the number of publications in this domain and its application to the real life of our hospitals. Finally, future orientations should include the cost assessment, the reproducibility of the results, and the usefulness “patient per patient” of liquid biopsy in comparison with current “solid biopsy” performance in lung cancer patients.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29. [PubMed]
- Gower A, Wang Y, Giaccone G. Oncogenic drivers, targeted therapies, and acquired resistance in non-small-cell lung cancer. J Mol Med (Berl) 2014;92:697-707. [PubMed]
- Holohan C, Van Schaeybroeck S, Longley DB, et al. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 2013;13:714-26. [PubMed]
- Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem 2013;59:110-8. [PubMed]
- Esposito A, Bardelli A, Criscitiello C, et al. Monitoring tumor-derived cell-free DNA in patients with solid tumors: clinical perspectives and research opportunities. Cancer Treat Rev 2014;40:648-55. [PubMed]
- Isobe K, Hata Y, Kobayashi K, et al. Clinical significance of circulating tumor cells and free DNA in non-small cell lung cancer. Anticancer Res 2012;32:3339-44. [PubMed]
- Taenzer A, Alix-Panabieres C, Wikman H, et al. Circulating tumor-derived biomarkers in lung cancer. J Thorac Dis 2012;4:448-9. [PubMed]
- Esmaeilsabzali H, Beischlag TV, Cox ME, et al. Detection and isolation of circulating tumor cells: principles and methods. Biotechnol Adv 2013;31:1063-84. [PubMed]
- Friedlander TW, Premasekharan G, Paris PL. Looking back, to the future of circulating tumor cells. Pharmacol Ther 2014;142:271-80. [PubMed]
- Han Y, Su C, Liu Z. Methods for detection of circulating cells in non-small cell lung cancer. Front Biosci (Landmark Ed) 2014;19:896-903. [PubMed]
- Liberko M, Kolostova K, Bobek V. Essentials of circulating tumor cells for clinical research and practice. Crit Rev Oncol Hematol 2013;88:338-56. [PubMed]
- Harouaka R, Kang Z, Zheng SY, et al. Circulating tumor cells: advances in isolation and analysis, and challenges for clinical applications. Pharmacol Ther 2014;141:209-21. [PubMed]
- Hofman V, Ilie M, Long E, et al. Detection of circulating tumor cells from lung cancer patients in the era of targeted therapy: promises, drawbacks and pitfalls. Curr Mol Med 2014;14:440-56. [PubMed]
- Millner LM, Linder MW, Valdes R Jr. Circulating tumor cells: a review of present methods and the need to identify heterogeneous phenotypes. Ann Clin Lab Sci 2013;43:295-304. [PubMed]
- Krebs MG, Metcalf RL, Carter L, et al. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol 2014;11:129-44. [PubMed]
- Gorges TM, Tinhofer I, Drosch M, et al. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012;12:178. [PubMed]
- Kim MY, Oskarsson T, Acharyya S, et al. Tumor self-seeding by circulating cancer cells. Cell 2009;139:1315-26. [PubMed]
- Hofman V, Bonnetaud C, Ilie MI, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res 2011;17:827-35. [PubMed]
- Hofman V, Ilie MI, Long E, et al. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay and the isolation by size of epithelial tumor cell method. Int J Cancer 2011;129:1651-60. [PubMed]
- Normanno N, Rossi A, Morabito A, et al. Prognostic value of circulating tumor cells’ reduction in patients with extensive small-cell lung cancer. Lung Cancer 2014;85:314-9. [PubMed]
- Gorges TM, Pantel K. Circulating tumor cells as therapy-related biomarkers in cancer patients. Cancer Immunol Immunother 2013;62:931-9. [PubMed]
- Ilie M, Long E, Butori C, et al. ALK-gene rearrangement, a comparative analysis on circulating tumour cells and tumour tissue from lung adenocarcinoma patients. Ann Oncol 2012;23:2907-13. [PubMed]
- Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366-77. [PubMed]
- Huang T, Jia CP, Jun Y, et al. Highly sensitive enumeration of circulating tumor cells in lung cancer patients using a size-based filtration microfluidic chip. Biosens Bioelectron 2014;51:213-8. [PubMed]
- Hofman VJ, Ilie MI, Bonnetaud C, et al. Cytopathologic detection of circulating tumor cells using the isolation by size of epithelial tumor cell method: promises and pitfalls. Am J Clin Pathol 2011;135:146-56. [PubMed]
- Hofman V, Long E, Ilie M, et al. Morphological analysis of circulating tumour cells in patients undergoing surgery for non-small cell lung carcinoma using the isolation by size of epithelial tumour cell (ISET) method. Cytopathology 2012;23:30-8. [PubMed]
- Yao X, Choudhury AD, Yamanaka YJ, et al. Functional analysis of single cells identifies a rare subset of circulating tumor cells with malignant traits. Integr Biol (Camb) 2014;6:388-98. [PubMed]
- Stroun M, Lyautey J, Lederrey C, et al. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta 2001;313:139-42. [PubMed]
- Sozzi G, Conte D, Leon M, et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol 2003;21:3902-8. [PubMed]
- Ulivi P, Mercatali L, Zoli W, et al. Serum free DNA and COX-2 mRNA expression in peripheral blood for lung cancer detection. Thorax 2008;63:843-4. [PubMed]
- Lee YJ, Yoon KA, Han JY, et al. Circulating cell-free DNA in plasma of never smokers with advanced lung adenocarcinoma receiving gefitinib or standard chemotherapy as first-line therapy. Clin Cancer Res 2011;17:5179-87. [PubMed]
- Paci M, Maramotti S, Bellesia E, et al. Circulating plasma DNA as diagnostic biomarker in non-small cell lung cancer. Lung Cancer 2009;64:92-7. [PubMed]
- Sozzi G, Roz L, Conte D, et al. Plasma DNA quantification in lung cancer computed tomography screening: five-year results of a prospective study. Am J Respir Crit Care Med 2009;179:69-74. [PubMed]
- Gautschi O, Bigosch C, Huegli B, et al. Circulating deoxyribonucleic Acid as prognostic marker in non-small-cell lung cancer patients undergoing chemotherapy. J Clin Oncol 2004;22:4157-64. [PubMed]
- Casoni GL, Ulivi P, Mercatali L, et al. Increased levels of free circulating DNA in patients with idiopathic pulmonary fibrosis. Int J Biol Markers 2010;25:229-35. [PubMed]
- Chang CP, Chia RH, Wu TL, et al. Elevated cell-free serum DNA detected in patients with myocardial infarction. Clin Chim Acta 2003;327:95-101. [PubMed]
- Rainer TH, Lam NY. Circulating nucleic acids and critical illness. Ann N Y Acad Sci 2006;1075:271-7. [PubMed]
- Jung K, Fleischhacker M, Rabien A. Cell-free DNA in the blood as a solid tumor biomarker--a critical appraisal of the literature. Clin Chim Acta 2010;411:1611-24. [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [PubMed]
- Nygaard AD, Holdgaard PC, Spindler KL, et al. The correlation between cell-free DNA and tumour burden was estimated by PET/CT in patients with advanced NSCLC. Br J Cancer 2014;110:363-8. [PubMed]
- Yu J, Gu G, Ju S. Recent advances in clinical applications of circulating cell-free DNA integrity. Lab Med 2014;45:6-11. [PubMed]
- Del Re M, Vasile E, Falcone A, et al. Molecular analysis of cell-free circulating DNA for the diagnosis of somatic mutations associated with resistance to tyrosine kinase inhibitors in non-small-cell lung cancer. Expert Rev Mol Diagn 2014;14:453-68. [PubMed]
- Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [PubMed]
- Devonshire AS, Whale AS, Gutteridge A, et al. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem 2014;406:6499-512. [PubMed]
- Pinzani P, Salvianti F, Pazzagli M, et al. Circulating nucleic acids in cancer and pregnancy. Methods 2010;50:302-7. [PubMed]
- Pinzani P, Salvianti F, Orlando C, et al. Circulating cell-free DNA in cancer. Methods Mol Biol 2014;1160:133-45. [PubMed]
- Vallée A, Marcq M, Bizieux A, et al. Plasma is a better source of tumor-derived circulating cell-free DNA than serum for the detection of EGFR alterations in lung tumor patients. Lung Cancer 2013;82:373-4. [PubMed]
- Xue X, Teare MD, Holen I, et al. Optimizing the yield and utility of circulating cell-free DNA from plasma and serum. Clin Chim Acta 2009;404:100-4. [PubMed]
- Tanaka T, Nagai Y, Miyazawa H, et al. Reliability of the peptide nucleic acid-locked nucleic acid polymerase chain reaction clamp-based test for epidermal growth factor receptor mutations integrated into the clinical practice for non-small cell lung cancers. Cancer Sci 2007;98:246-52. [PubMed]
- Kimura H, Fujiwara Y, Sone T, et al. High sensitivity detection of epidermal growth factor receptor mutations in the pleural effusion of non-small cell lung cancer patients. Cancer Sci 2006;97:642-8. [PubMed]
- Couraud S, Vaca-Paniagua F, Villar S, et al. Non-invasive diagnosis of actionable mutations by deep sequencing of circulating-free DNA in non-small cell lung cancer: Findings from BioCAST / IFCT-1002. Clin Cancer Res 2014;20:4613-24. [PubMed]
- Pailler E, Adam J, Barthelemy A, et al. Detection of Circulating Tumor Cells Harboring a Unique ALK Rearrangement in ALK-Positive Non-Small-Cell Lung Cancer. J Clin Oncol 2013;31:2273-81. [PubMed]
- Dziadziuszko R, Wynes MW, Singh S, et al. Correlation between MET gene copy number by silver in situ hybridization and protein expression by immunohistochemistry in non-small cell lung cancer. J Thorac Oncol 2012;7:340-7. [PubMed]
- Hofman P, Ilie M, Hofman V, et al. Immunohistochemistry to identify EGFR mutations or ALK rearrangements in patients with lung adenocarcinoma. Ann Oncol 2012;23:1738-43. [PubMed]
- Hofman V, Ilie M, Long-Mira E, et al. Usefulness of immunocytochemistry for the detection of the BRAF(V600E) mutation in circulating tumor cells from metastatic melanoma patients. J Invest Dermatol 2013;133:1378-81. [PubMed]
- Ilie MI, Hofman V, Bonnetaud C, et al. Usefulness of tissue microarrays for assessment of protein expression, gene copy number and mutational status of EGFR in lung adenocarcinoma. Virchows Arch 2010;457:483-95. [PubMed]
- Yoshida A, Tsuta K, Wakai S, et al. Immunohistochemical detection of ROS1 is useful for identifying ROS1 rearrangements in lung cancers. Mod Pathol 2014;27:711-20. [PubMed]
- Benesova L, Belsanova B, Suchanek S, et al. Mutation-based detection and monitoring of cell-free tumor DNA in peripheral blood of cancer patients. Anal Biochem 2013;433:227-34. [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [PubMed]
- Rosell R, Molina MA, Serrano MJ. EGFR mutations in circulating tumour DNA. Lancet Oncol 2012;13:971-3. [PubMed]
- Taniguchi K, Uchida J, Nishino K, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 2011;17:7808-15. [PubMed]
- Nygaard AD, Garm Spindler KL, Pallisgaard N, et al. The prognostic value of KRAS mutated plasma DNA in advanced non-small cell lung cancer. Lung Cancer 2013;79:312-7. [PubMed]
- El Messaoudi S, Rolet F, Mouliere F, et al. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta 2013;424:222-30. [PubMed]
- Page K, Guttery DS, Zahra N, et al. Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS One 2013;8:e77963. [PubMed]
- Vallée A, Le Loupp AG, Denis MG. Efficiency of the Therascreen(R) RGQ PCR kit for the detection of EGFR mutations in non-small cell lung carcinomas. Clin Chim Acta 2014;429:8-11. [PubMed]
- Adalsteinsson VA, Love JC. Towards Engineered Processes for Sequencing-Based Analysis of Single Circulating Tumor Cells. Curr Opin Chem Eng 2014;4:97-104. [PubMed]
- Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4:136ra68.
- Hodgkinson CL, Morrow CJ, Li Y, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 2014;20:897-903. [PubMed]
- Kukita Y, Uchida J, Oba S, et al. Quantitative identification of mutant alleles derived from lung cancer in plasma cell-free DNA via anomaly detection using deep sequencing data. PLoS One 2013;8:e81468. [PubMed]
- Ni X, Zhuo M, Su Z, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A 2013;110:21083-8. [PubMed]
- Ilie M, Long E, Hofman V, et al. Clinical value of circulating endothelial cells and of soluble CD146 levels in patients undergoing surgery for non-small cell lung cancer. Br J Cancer 2014;110:1236-43. [PubMed]
- Sanfiorenzo C, Ilie MI, Belaid A, et al. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One 2013;8:e54596. [PubMed]


