
CircRNA ZNF609 in peripheral blood leukocytes acts as a protective factor and a potential biomarker for coronary artery disease
Introduction
Coronary artery disease (CAD) is a complex and multifactorial disease and remains the leading cause of death worldwide (1). In CAD pathogenesis theories, the inflammatory response theory has been widely recognized (2). Lately, the COLCOT trials including 4,745 myocardial infarction participants reported that anti-inflammatory medication colchicine significantly reduced the risk of ischemic cardiovascular events (3). Besides, the CANTOS trial enrolling 10,061 high-risk patients defined by prior myocardial infarction has demonstrated that canakinumab targeting the IL-1β anti-inflammatory pathway significantly reduced the incidence of cardiovascular events (4). These two breakthrough clinical studies have lent strong support to the theory that inflammation contributes to the pathogenesis of atherosclerotic diseases. Considered that inflammation plays a crucial role in the atherosclerosis process (5), it is feasible to explore potential inflammation-related biomarkers for prediction and surveillance of cardiovascular disease.
Circular RNA (circRNA), a novel class of endogenous noncoding RNAs, is quickly gaining prominence rapidly due to the discovery of its regulatory role and predictive value in several pathological processes (6-9). Because of their covalently closed loop structure and resistance to nuclease, circRNAs have become convenient and ideal molecular predictive biomarkers (10-12). CircRNAs have been found to be involved in several cardiovascular pathological processes, including myocardial infarction (13), heart failure (14) and overall atherosclerosis (15). CircRNA ZNF609 (circBase ID: hsa_circ_0000615) is a circle RNA formed by the 8th-10th exons of the host gene ZNF609 (Zinc Finger family 609), which located at chr15:64791491-64792365. Liu et al. (16) reported that circZNF609 could participate in the regulation of vascular endothelial function by interacting with miR-615-5p. Studies have shown that zinc finger proteins play important roles in inflammatory regulation (17), apoptosis regulation (18), and lipid metabolism (19). Since inflammation (20), apoptosis (21), lipid metabolism are all involved in the CAD and vascular endothelial dysfunction is considered of paramount importance in the atherosclerotic process (22), we hypothesized that circZNF609 could be related to CAD pathogenesis (16).
Given present data, we carried out this study to investigate the role of circZNF609 in peripheral blood leukocytes (PBLs) of CAD patients, and to evaluate its possible diagnostic value as a novel biomarker. We present the following article in accordance with the STARD reporting checklist (available at http://dx.doi.org/10.21037/ atm-19-4728).
Methods
Experimental subjects
A total of 330 CAD patients and 209 controls were recruited from Zhongnan Hospital of Wuhan University between April 2016 and May 2017. Of these, 30 control individuals and 30 CAD patients with different Gensini scores were randomly selected as the discovery cohort, whereas the remaining population served as the validation group. The diagnostic criteria of CAD were defined as stenosis >50% in at least one of the coronary arteries by invasive coronary angiography (ICA). CAD was used to be classified into four subtypes of stable angina pectoris (SAP), unstable angina pectoris (UA), non-ST-segment elevation myocardial infarction (NSTEMI) and ST-segment elevation myocardial infarction (STEMI), while the latest 2019 ESC Guidelines on Chronic Coronary Syndromes emphasized that CAD was classified into acute coronary syndromes (ACS) and chronic coronary syndromes (CCS) (23). Patients with the following diseases were excluded: (I) cardiac diseases including congenital heart diseases, myocardial bridge, cardiomyopathy or severe non-coronary cardiovascular diseases; (II) systemic acute or chronic infections or inflammatory diseases; (III) malignant tumors; (IV) autoimmune diseases; (V) liver and kidney dysfunctions; (VI) endocrine diseases such as thyroid diseases. Controls were age- and gender-matched individuals who came from the Physical Examination Center without cardiac diseases or any of the aforementioned conditions.
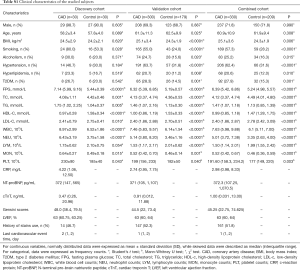
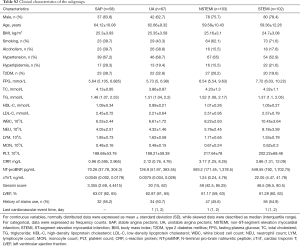
The extracted data included demographic information: age, gender, body mass index (BMI) and traditional CAD risk factors: smoking, alcohol drinking, histories of hypertension, dyslipidemia and type 2 diabetes mellitus (T2DM). Fasting plasma glucose (FPG), total cholesterol (TC), total triglyceride (TG), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), routine complete blood count, C-reaction protein (CRP), N-terminal pro–brain natriuretic peptide (NT-proBNP), and cardiac troponin T (cTnT) were measured by standard techniques in the Clinical Core Laboratory of Zhongnan Hospital. Detailed information is depicted in Tables S1,S2.
Full table
Full table
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Medical Institutional Review Board of Zhongnan Hospital of Wuhan University (approval number 2010052). Informed consent was obtained before the study.
Calculation of the Gensini score
The severity of coronary atherosclerosis was assessed by the Gensini score, which took the extent and the location of the stenosis into account (24). The 0–25%, 26–50%, 51–75%, 76–90%, 91–99% and 100% stenosis were scored at 1, 2, 4, 8, 16 and 32 respectively. Different coronary artery branches corresponded to different coefficients, ranging from 0.5 to 5, depending on the stenosis location and significance of the cardiac region supplied by that segment. Gensini scores of each patient were sum of scores of coronary artery branches multiplied by the corresponding coefficients.
Calculation of monocyte-to-lymphocyte ratio (MLR), platelet-to-lymphocyte ratio (PLR), neutrophil-to-lymphocyte ratio (NLR)
The MLR, PLR and NLR were calculated based on the counts of neutrophils, lymphocytes, monocytes, and platelets. MLR = monocyte counts/lymphocyte counts, PLR = platelet counts/lymphocyte counts, NLR = neutrophil counts/lymphocyte counts.
Cell culture
RAW264.7 cells were suspended in DMEM medium (Gibco, USA) containing 10% fetal bovine serum (Gibco, USA) and seeded in 6-well culture plates at a concentration of 4.0×105 cells/well. At 60–80% confluence, pcDNA3.1-circZNF609 vector or the blank control vector (GenePharma, China) were transfected with FuGENE HD transfection reagent (Promega, USA) at a ratio of 2 µg DNA/7 µL FuGENE (according to the manufacturer’s recommendations). After 24 h of transfection, cells were collected for subsequent experiments.
RNA extraction and quantitative real time polymerase chain reaction
Fresh whole blood was centrifuged at 3,000 rpm at 4 °C for 10 min to separate blood cells and plasma. The blood cells were added to the erythrocyte lysate at a ratio of 1:3 and incubated at 4 °C until the erythrocytes were completely ruptured. After 12,000 rpm centrifugation at 4 °C for 5 min, the supernatant was discharged and the precipitation was PBLs. Total RNA in PBLs and RAW264.7 were isolated using TRIzol reagent (Invitrogen, USA). Nanodrop 2000 (Thermo Scientific, USA) was used to detect the concentration and purity of RNA. Total RNA (2 µg) was incubated with or without 3 U/µg of RNase R (Epicentre Technologies, USA) for 30 min at 37 °C, and cDNA was synthesized according to the manufacturer instructions of PrimeScriptTM RT reagent Kit (Toyobo, Japan).
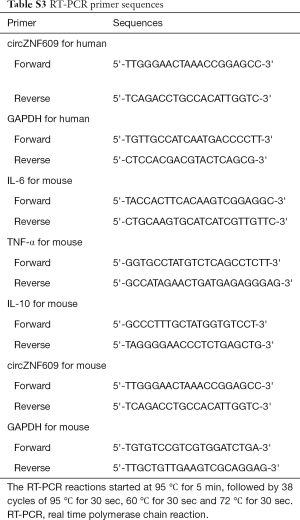
The mRNA expression levels of circZNF609, IL-6, TNF-α, IL-10 were detected using quantitative real time polymerase chain reaction (RT-PCR) on the Bio-Rad CFX96 (Bio-Rad, USA) with the Fast SYBR® Green PCR Master Mix-PE (Applied Biosystem, USA). Relative expressions were calculated as 2−ΔCt using GAPDH as a reference gene. Primers are listed in Table S3.
Full table
Prediction for downstream pathways of circZNF609
To investigate the underlying mechanisms of expression variations of circZNF609 in CAD, TargetScan (25) was used to predict target microRNAs of circZNF609. DIANA-miRPath v.3 platform (TarBase v7.0 method) (26) was used for pathway enrichment analysis of microRNAs. Then, we retrieved four complementary microRNAs with circZNF609 on PubMed, which were confirmed by previous reported double luciferase assays. The potential targets of these microRNAs were predicted by MiRWalk (27). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were carried out based on DAVID Bioinformatics Resources 6.8 (28). circZNF609-miRNAs-mRNA network map was drawn by Cytoscape Software (v3.7.1) (29).
Statistical analysis
Data normally distributed were presented as mean ± standard deviation (SD), otherwise being described by the median with inter-quartile range. Normality of data distribution was assessed by the Shapiro-Wilk test. The normally distributed continuous variables were assessed by the Student’s t-test, while non-normally distributed variables were analyzed by the Mann-Whitney test. Categorical variables were analyzed by the chi-square test. When comparison was performed among multiplied groups, One-way ANOVA test or nonparametric test was used. The Spearman correlation and multivariate regression were used to test correlations between circZNF609 expression levels and clinical characteristics in CAD patients. receiver operating characteristic curve (ROC curve) analysis was conducted to estimate the potential clinical predictive values. Associations between circZNF609 relative expressions and CAD were analyzed by the multivariate logistic regression and unconditional logistic regression. P<0.05 was considered to be statistically significant. All statistical analyses were performed on SPSS version 23.0 (SPSS, USA) and GraphPad Prism 8.0 (GraphPad Software, USA).
Results
Clinical characteristics in the combined cohort
The clinical characteristics are depicted in Table S1. There were no differences in age or gender between CAD and control groups in the combined cohort. The BMI level (25.1±3.6 vs. 24.3±1.9, P=0.006), smoking status (57.3% vs. 28.2%, P<0.0001), alcohol drinking (25.2% vs. 16.3%, P=0.015), hypertension (62.4% vs. 31.6%, P<0.0001), hyperlipidemia (20.6% vs. 12.0%, P=0.010) and T2DM (27.9% vs. 15.3%, P=0.001) in the CAD group were significantly higher than the control group.
Specific detection of circZNF609 expressions
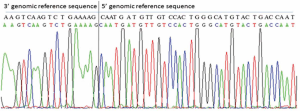
To verify the specificity of the primers for circZNF609 amplification, RT-PCR products were sequenced and compared with the standard sequence of circZNF609 in the circBase database. The Sanger sequencing results showed that the product sequence was consistent with the database sequence, suggesting that circZNF609 could be specifically amplified by RT-PCR (Figure S1).
Decreased circZNF609 expression levels in CAD patients
In the discovery group, the expression level of circZNF609 was significantly decreased in PBLs of patients, compared with controls (P<0.0001) (Figure 1A). Furthermore, we validated the decrease circZNF609 in the validation cohort consisting of 179 controls and 300 CAD patients (Figure 1B, P<0.0001). In the combined cohort, circZNF609 was still significantly down-regulated in the CAD group (P<0.0001) (Figure 1C). In addition, circZNF609 was statistically lower in non-T2DM CAD patients compared with non-T2DM controls, while there was no difference between CAD patients with vs. without T2DM (Figure S2). These data were coincident with the logistic regression analysis results.


The expression of circZNF609 was an independent protective factor for CAD
Without adjustment, the OR for CAD was 0.056 (95% CI: 0.270–0.114, P<0.0001) in individuals with the expression of circZNF609 in the top tertile compared with the bottom tertile. After progressive adjustment for age, gender, BMI, smoking, alcoholism, histories of hypertension, hyperlipidemia, T2DM, history of statins use, PBLs counts and PBLs classification, the expression of circZNF609 remained significantly associated with CAD prevalence (Table 1).

Full table
The expression of circZNF609 was negatively related to CRP levels in cases
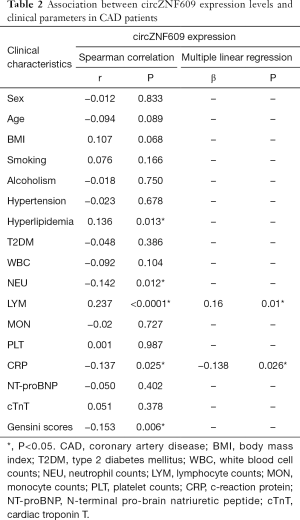
The results of the Spearman correlation analysis showed that the expression level of circZNF609 was negatively correlated with CRP levels (r=−0.137, P=0.025), neutrophils counts (r=−0.142, P=0.012) and Gensini scores (r=−0.153, P=0.006), and positively correlated with lymphocyte counts (r=0.237, P<0.0001) and history of hyperlipidemia (r=0.136, P=0.013). After adjustment for several differential clinical parameters in stepwise multivariate linear regression model, we found that lymphocyte counts (β=0.16, P=0.01) and CRP (β=−0.138, P=0.026) were independent factors associated with circZNF609 expressions (Table 2). Since the related indexes were inflammatory factor, we further analyzed the relationship between circZNF609 expression and the systemic inflammatory index (SII), and found circZNF609 was negatively correlated with MLR (r=−0.168, P=0.002), NLR (r=−0.221, P<0.0001) and PLR (r=−0.158, P=0.004) (Figure S3). These results suggest that circZNF609 may be closely related to inflammatory processes in CAD.
Full table
Diagnostic values of circZNF609 expressions in PBLs.
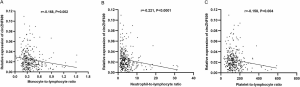
ROC was constructed to assess whether circZNF609 expressions could be used as a potential diagnostic marker for CAD. In the discovery cohort, the AUC was 0.830 (95% CI: 0.726–0.925, P<0.0001) (Figure 2A), while the AUC of the validation cohort was 0.752 (95% CI: 0.710–0.800, P<0.0001) (Figure 2B). Overall, the AUC, specificity and sensitivity for CAD of circZNF609 were 0.761 (95% CI: 0.721–0.800, P<0.0001), 0.804 (95% CI: 0.745–0.852) and 0.615 (95% CI: 0.562–0.666), respectively. A circZNF609 expression level of 0.024 was the best cut-off value (30). These results indicated a moderate predictive value of circZNF609 for distinguishing patients with CAD from controls (Figure 2C).
The expression level of circZNF609 in different clinical subgroups.
The expression levels of circZNF609 among four CAD subgroups: SAP (n=58), UA (n=67), NSTEMI (n=103) and STEMI (n=102) were different but all significantly decreased compared to the control group (all P<0.0001) (Figure 3A). The AUC of circZNF609 was 0.705 (95% CI: 0.624–0.787, P<0.0001) in SAP, 0.807 (95% CI: 0.740–0.873, P<0.0001) in UA, 0.713 (95% CI: 0.648–0.778, P<0.0001) in NSTEMI, and 0.752 (95% CI: 0.691–0.813, P<0.0001) in STEMI (Figure 3B). The corresponding specificity and sensitivity of each subtype are presented in Table 3.


Full table
Overexpression of circZNF609 attenuated inflammation
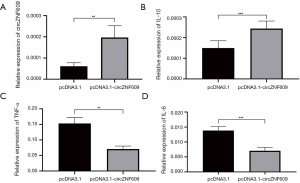
In order to further explore the potential function of circZNF609 on the inflammatory response, pcDNA3.1-circZNF609 vector or the blank pcDNA3.1 vector were transfected into RAW264.7 cells. The results showed the pro-inflammatory cytokines including IL-6 and TNF-α in cells transfected with pcDNA3.1-circZNF609 was decreased compared with the control vector, whereas the anti-inflammatory cytokine IL-10 was significantly increased (Figure 4), which indicated that circZNF609 might play a protective role in the inflammatory response of CAD by modulating inflammation cytokines.
Annotation for circZNF609 downstream pathways
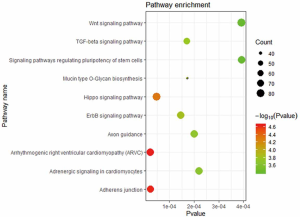
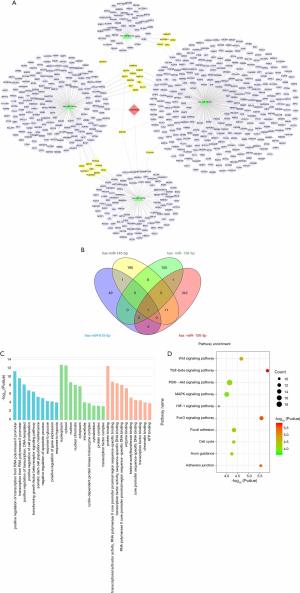
To examine the potential downstream pathway of circZNF609, KEGG pathway enrichment analysis was performed on the miRNAs predicted by TargetScan interacting with circZNF609 (Figure S4), and results showed they were mainly enrichment in Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC), adherens junction, TGF-β signaling pathway, etc., which were highly related to the pathophysiological process of CAD. Then, we found four complementary microRNAs with circZNF609 on PubMed, hsa-miR-145-5p (31), hsa-miR-615-5p (16), hsa-miR-138-5p (32), hsa-miR-150-5p (33), which were confirmed by double luciferase assays in previous reports. We selected validated target genes from the predicted targets results of MiRWalk as downstream targets (Table S4). A network map comprising circZNF609, four miRNAs and their target genes was presented in Figure 5A. Venn diagram revealed the number of common downstream targets of the four miRNAs (Figure 5B). GO and KEGG pathway analysis of those target genes showed that they were mainly concentrated in the pathway closely related to CAD, in which the pro-inflammatory factors IL-6 and TNF-ɑ were pivotal irritants of HIF-1α and MAPK signaling pathways, respectively (Figure 5C,D).

Full table
Discussion
In the present study, we found that circZNF609 was significantly decreased in PBLs of CAD patients and might be an independent protective factor for CAD (Table 1). Furthermore, we identified its’ anti-inflammation function, based on the independent negative correlation of circZNF609 with systemic inflammation indexes (Table 2) and in vitro experiments results (Figure 4). In addition, it could be a potential diagnostic biomarker for CAD with the diagnostic AUC value of 0.761.
So far, the theory that chronic inflammatory processes persist in the course of CAD has been gaining the spotlight (34). CRP has been shown to be a marker of systemic inflammation (35), while elevated circulating levels of highly sensitive CRP (hsCRP) are strongly associated with future vascular events, independent of established risk factors (36,37). We found circZNF609 was reversely correlated with the systemic inflammation indexes, CRP and MLR, NLR and PLR, which have been proven to be closely related to the occurrence, development and prognosis of CAD (38-41). MLR and NLR were the independent predictors of cardiovascular diseases and positively associated with the extent and severity of coronary lesions (42,43). Increased PLR might be a marker of chronic inflammation, which would ultimately lead to increased CAD risk factors such as slow coronary blood flow and platelet-monocyte aggregation formation (44,45). Furthermore, MLR, NLR and PLR were reported to be positively correlated with IL-6 (46), TNF-ɑ (47) and CRP (48) respectively, while circZNF609 was negatively correlated with these systemic inflammation indexes in our study. These supported that circZNF609 might play a protective function in CAD through an anti-inflammatory pathway.
Recently, circRNAs were proven to have the capacity to regulate the inflammatory process of atherosclerosis. The A20 gene in the zinc finger protein family was commonly deemed as an anti-inflammatory and nuclear factor-kappa B (NF-κB) inhibitor that leads to decreased expression of the downstream pro-inflammatory cytokine IL-6 and TNF-α by inhibiting the NF-κB signaling pathway (49). We guessed whether circZNF609 have similar functions with A20. After overexpression of circZNF609 in RAW264.7 cells, it was found that the expression of IL-6 and TNF-α were significantly decreased, while the expression of anti-inflammatory cytokines IL-10 was increased. The results confirmed that circZNF609 might function in CAD through reducing the intensity of inflammatory response by decreasing pro-inflammatory cytokines and increasing anti-inflammatory cytokines.
There are growing evidences supporting the added value of circRNA in the diagnosis of CAD, since circRNA is more stable than linear RNA because of the covalently closed loop structures and resistance to RNA exonuclease (50,51). Burd et al. found that the circular isoform of ANRIL (cANRIL) may affect the progression of CAD by regulating INK4/ARF expression (52). Wang et al. found that the overexpression of hsa_circ_0004104 in THP-1-derived macrophages induced the up-regulation of pro-atherosclerotic genes, which implying that circRNA might be involved in the pathologies of CAD (53). In our study, the sensitivity and specificity of circZNF609 for the CAD diagnosis were 0.615 and 0.804, respectively, which were greater than routine ECG [sensitivity and specificity were 0.29 and 0.67, respectively (54)]. Considering the cost and convenience of diagnostic methods, circZNF609 might assist in the lab diagnosis of CAD.
In the present study, we divided CAD into four subtypes according to previous clinical classifications, though the latest 2019 ESC Guidelines on Chronic Coronary Syndromes classified CAD into ACS and CCS. Though the new classification could better reflect the pathophysiological characteristics of the dynamic changes in CAD, we think the previous classification classified the patients from the aspect of chronic inflammation process in CAD. SAP and UA could mainly be the inflammation of vessels, while NSTEMI and STEMI involved the vessel and cardiac inflammation which might induce an enhanced inflammatory reaction. It could probably be the reason that there was the lowest circZNF609 level in UA with a highest diagnostic AUC value, but a climbing in NSTEMI and STEMI. We inferred that there might be other confounding factors (for example cytokines releasing by myocardial necrosis etc.) up-regulating circZNF609 in MI.
Recent studies have reported that circRNAs acted as miRNA sponges to relieve the inhibitory effects of a miRNA on its target genes, thereby increasing the expression of the target genes (55). In this study, the potential pathway of circZNF609 was explored by bioinformatics databases, and we further predicted the four miRNAs’ targets, which are mainly concentrated in the signaling pathways related to inflammation. AKT1 was predicted to be the targets of miR-138-5p, and it was proven that IL-6 would increase if AKT1 was silencing in mouse macrophage RAW264.7 (56). AKT1 exerts inhibitory effect in the MAPK signaling pathway through KEGG (https://www.kegg.jp/). Moreover, miR-138-5p was reported to retain SRY-related high-mobility-group-box 9 (SOX9) to facilitate the inflammation, which is a well-acknowledged transcription factor (57). In addition, inhibiting miR‐150‐5p alleviates cardiac inflammation via targeting Smad7 in cardiac fibroblasts (58). Based on the bioinformatics prediction and the literature reports, we speculated that circZNF609 sponge hsa-miR-138-5p and hsa-miR-150-5p to enhance anti-inflammation genes and to interrupter inflammation pathways to alleviate the atherosclerosis chronic inflammation.
It should be noted that there were some limitations in our study. A larger sample size and multi-center study is needed to further verify our results. Due to the lack of follow-up information for CAD patients, the prognostic values of circZNF609 in major cardiovascular events in apparent normal population should be evaluated in subsequent studies. The role of circZNF609 in CAD disease causal pathways was not fully demonstrated in our study, although this knowledge is not a prerequisite for biomarkers.
In summary, we reported a downregulated circZNF609 in PBLs of CAD patients, which might function as an independent protective factor of CAD through the anti-inflammation pathway, and could be used as a new biomarker for the diagnosis of CAD. In future studies, we will focus on the specific pronounced function of circZNF609 in the pathological process of CAD.
Acknowledgments
Funding: This study was supported by the Grant of National Natural Science Foundation of China (Grant No. 81871722).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/atm-19-4728
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-19-4728
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-19-4728). All authors report grants from the Grant of National Natural Science Foundation of China, during the conduct of the study; in addition, BL, ML, YX and FZ have a patent—The primer design and clinical application of a nucleic acid biomarker for CAD pending.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Medical Institutional Review Board of Zhongnan Hospital of Wuhan University (approval number 2010052). Informed consent was obtained before the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dagenais GR, Leong DP, Rangarajan S, et al. Variations in common diseases, hospital admissions, and deaths in middle-aged adults in 21 countries from five continents (PURE): a prospective cohort study. Lancet 2020;395:785-94. [Crossref] [PubMed]
- Ross R. Atherosclerosis--an inflammatory disease. N Engl J Med 1999;340:115-26. [Crossref] [PubMed]
- Tardif JC, Kouz S, Waters DD, et al. Efficacy and Safety of Low-Dose Colchicine after Myocardial Infarction. N Engl J Med 2019;381:2497-505. [Crossref] [PubMed]
- Ridker PM, Everett BM, Thuren T, et al. Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N Engl J Med 2017;377:1119-31. [Crossref] [PubMed]
- Geovanini GR, Libby P. Atherosclerosis and inflammation: overview and updates. Clin Sci (Lond) 2018;132:1243-52. [Crossref] [PubMed]
- Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 2017;66:1151-64. [Crossref] [PubMed]
- Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer 2018;17:19. [Crossref] [PubMed]
- Li LJ, Huang Q, Pan HF, et al. Circular RNAs and systemic lupus erythematosus. Exp Cell Res 2016;346:248-54. [Crossref] [PubMed]
- Li TR, Jia YJ, Wang Q, et al. Circular RNA: a new star in neurological diseases. Int J Neurosci 2017;127:726-34. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792-806. [Crossref] [PubMed]
- Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015;160:1125-34. [Crossref] [PubMed]
- Geng HH, Li R, Su YM, et al. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PLoS One 2016;11:e0151753. [Crossref] [PubMed]
- Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016;37:2602-11. [Crossref] [PubMed]
- Shen L, Hu Y, Lou J, et al. CircRNA0044073 is upregulated in atherosclerosis and increases the proliferation and invasion of cells by targeting miR107. Mol Med Rep 2019;19:3923-32. [PubMed]
- Liu C, Yao MD, Li CP, et al. Silencing Of Circular RNA-ZNF609 Ameliorates Vascular Endothelial Dysfunction. Theranostics 2017;7:2863-77. [Crossref] [PubMed]
- Zou Q, Gang K, Yang Q, et al. The CCCH-type zinc finger transcription factor Zc3h8 represses NF-kappaB-mediated inflammation in digestive organs in zebrafish. J Biol Chem 2018;293:11971-83. [Crossref] [PubMed]
- Tian C, Xing G, Xie P, et al. KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol 2009;11:580-91. [Crossref] [PubMed]
- Wagner S, Hess MA, Ormonde-Hanson P, et al. A broad role for the zinc finger protein ZNF202 in human lipid metabolism. J Biol Chem 2000;275:15685-90. [Crossref] [PubMed]
- Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med 2005;352:1685-95. [Crossref] [PubMed]
- Bennett MR, Evan GI, Schwartz SM. Apoptosis of human vascular smooth muscle cells derived from normal vessels and coronary atherosclerotic plaques. J Clin Invest 1995;95:2266-74. [Crossref] [PubMed]
- Gutiérrez E, Flammer AJ, Lerman LO, et al. Endothelial dysfunction over the course of coronary artery disease. Eur Heart J 2013;34:3175-81. [Crossref] [PubMed]
- Knuuti J, Wijns W, Saraste A, et al. 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromes. Eur Heart J 2020;41:407-77. [Crossref] [PubMed]
- Gensini GG. A more meaningful scoring system for determining the severity of coronary heart disease. Am J Cardiol 1983;51:606. [Crossref] [PubMed]
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005. [Crossref] [PubMed]
- Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 2015;43:W460-6. [Crossref] [PubMed]
- Sticht C, De La Torre C, Parveen A, et al. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One 2018;13:e0206239. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 1993;39:561-77. [Crossref] [PubMed]
- Wang S, Xue X, Wang R, et al. CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6K1 via sponging miR-145-5p. Cancer Manag Res 2018;10:3881-90. [Crossref] [PubMed]
- Xiong Y, Zhang J, Song C. CircRNA ZNF609 functions as a competitive endogenous RNA to regulate FOXP4 expression by sponging miR-138-5p in renal carcinoma. J Cell Physiol 2019;234:10646-54. [Crossref] [PubMed]
- Zhu L, Liu Y, Yang Y, et al. CircRNA ZNF609 promotes growth and metastasis of nasopharyngeal carcinoma by competing with microRNA-150-5p. Eur Rev Med Pharmacol Sci 2019;23:2817-26. [PubMed]
- Hansson GK. Inflammation and Atherosclerosis: The End of a Controversy. Circulation 2017;136:1875-7. [Crossref] [PubMed]
- Deodhar SD. C-reactive protein: the best laboratory indicator available for monitoring disease activity. Cleve Clin J Med 1989;56:126-30. [Crossref] [PubMed]
- Ridker PM, Cushman M, Stampfer MJ, et al. Inflammation, aspirin, and the risk of cardiovascular disease in apparently healthy men. N Engl J Med 1997;336:973-9. [Crossref] [PubMed]
- Ridker PM, Hennekens CH, Buring JE, et al. C-reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. N Engl J Med 2000;342:836-43. [Crossref] [PubMed]
- Buckley DI, Fu R, Freeman M, et al. C-reactive protein as a risk factor for coronary heart disease: a systematic review and meta-analyses for the U.S. Preventive Services Task Force. Ann Intern Med 2009;151:483-95. [Crossref] [PubMed]
- Massiot N, Lareyre F, Voury-Pons A, et al. High Neutrophil to Lymphocyte Ratio and Platelet to Lymphocyte Ratio are Associated with Symptomatic Internal Carotid Artery Stenosis. J Stroke Cerebrovasc Dis 2019;28:76-83. [Crossref] [PubMed]
- Tamhane UU, Aneja S, Montgomery D, et al. Association between admission neutrophil to lymphocyte ratio and outcomes in patients with acute coronary syndrome. Am J Cardiol 2008;102:653-7. [Crossref] [PubMed]
- Lareyre F, Carboni J, Chikande J, et al. Association of Platelet to Lymphocyte Ratio and Risk of 30-Day Postoperative Complications in Patients Undergoing Abdominal Aortic Surgical Repair. Vasc Endovascular Surg 2019;53:5-11. [Crossref] [PubMed]
- Ji H, Li Y, Fan Z, et al. Monocyte/lymphocyte ratio predicts the severity of coronary artery disease: a syntax score assessment. BMC Cardiovasc Disord 2017;17:90. [Crossref] [PubMed]
- Verdoia M, Barbieri L, Di Giovine G, et al. Neutrophil to Lymphocyte Ratio and the Extent of Coronary Artery Disease: Results From a Large Cohort Study. Angiology 2016;67:75-82. [Crossref] [PubMed]
- Akboga MK, Canpolat U, Balci KG, et al. Increased Platelet to Lymphocyte Ratio is Related to Slow Coronary Flow. Angiology 2016;67:21-6. [Crossref] [PubMed]
- Ugur M, Gul M, Bozbay M, et al. The relationship between platelet to lymphocyte ratio and the clinical outcomes in ST elevation myocardial infarction underwent primary coronary intervention. Blood Coagul Fibrinolysis 2014;25:806-11. [Crossref] [PubMed]
- Serfőző G, Horvath T, Foldesi I, et al. The Monocyte-to-Lymphocyte Ratio Correlates with Psycho-Neuro-Inflammatory Factors in Patients with Stable Coronary Artery Disease. Neuroimmunomodulation 2016;23:67-74. [Crossref] [PubMed]
- Suárez-Cuenca JA, Ruiz-Hernandez AS, Mendoza-Castaneda AA, et al. Neutrophil-to-lymphocyte ratio and its relation with pro-inflammatory mediators, visceral adiposity and carotid intima-media thickness in population with obesity. Eur J Clin Invest 2019;49:e13085. [Crossref] [PubMed]
- Pan L, Du J, Li T, et al. Platelet-to-lymphocyte ratio and neutrophil-to-lymphocyte ratio associated with disease activity in patients with Takayasu's arteritis: a case-control study. BMJ Open 2017;7:e014451. [Crossref] [PubMed]
- Jarosz M, Olbert M, Wyszogrodzka G, et al. Antioxidant and anti-inflammatory effects of zinc. Zinc-dependent NF-kappaB signaling. Inflammopharmacology 2017;25:11-24. [Crossref] [PubMed]
- Memczak S, Papavasileiou P, Peters O, et al. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS One 2015;10:e0141214. [Crossref] [PubMed]
- Beermann J, Piccoli MT, Viereck J, et al. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev 2016;96:1297-325. [Crossref] [PubMed]
- Burd CE, Jeck WR, Liu Y, et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010;6:e1001233. [Crossref] [PubMed]
- Wang L, Shen C, Wang Y, et al. Identification of circular RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis 2019;286:88-96. [Crossref] [PubMed]
- Ghadrdoost B, Haghjoo M, Firouzi A. Accuracy of cardiogoniometry compared with electrocardiography in the diagnosis of coronary artery disease. Res Cardiovasc Med 2015;4:e25547. [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Tang B, Li X, Ren Y, et al. MicroRNA-29a regulates lipopolysaccharide (LPS)-induced inflammatory responses in murine macrophages through the Akt1/ NF-kappaB pathway. Exp Cell Res 2017;360:74-80. [Crossref] [PubMed]
- Chunlei H, Chang Z, Sheng L, et al. Down-regulation of MiR-138-5p Protects Chondrocytes ATDC5 and CHON-001 from IL-1 beta-induced Inflammation Via Up-regulating SOX9. Curr Pharm Des 2020;25:4613-21. [Crossref] [PubMed]
- Che H, Wang Y, Li Y, et al. Inhibition of microRNA-150-5p alleviates cardiac inflammation and fibrosis via targeting Smad7 in high glucose-treated cardiac fibroblasts. J Cell Physiol 2019. [Epub ahead of print]. [Crossref] [PubMed]


