
Treatment response in triple-negative breast cancer: a role for eEF2 kinase-mediated autophagy?
The intrinsic or acquired ability of cancers to survive clinical therapies remains one of the greatest obstacles in oncology. This ability is attributed to the dynamic nature of cancers, which facilitates the creation of heterogenous populations of cells that harbor unique molecular signatures within a tumor. Intrinsic treatment resistance predates clinical intervention and is a result of pre-existing genetic and molecular alterations that confer a survival advantage to some cancer cells. In contrast, acquired treatment resistance is a consequence of the development of new mutations and adaptations by subpopulations of cancer cells within a tumor that ultimately desensitize them to ongoing treatments (1). There is currently a large body of ongoing research dedicated towards dissecting and understanding the genetic and non-genetic alterations and pathways that contribute to the sensitivity of cancers to current clinical therapies (2). One cellular pathway that has been extensively studied in this regard over the past decade is macroautophagy (hereafter referred to as autophagy) (3,4).
Autophagy has the ability to suppress or support tumor initiation and progression, and this duality in function is largely dependent on the stage of tumorigenesis. Autophagy functions as a tumor suppressive pathway during early stages of tumorigenesis but supports tumor survival during later stages of cancer progression (5,6). The pro-survival roles of autophagy in cancer progression have primed it as an attractive therapeutic target for augmenting the effects of chemotherapies and targeted agents, and for overcoming treatment resistance. Autophagy consists of multiple sequential steps, and these steps present potentially druggable targets that have been leveraged to suppress autophagy in cancers (5). Late stage autophagy inhibitors that impair lysosome function, like chloroquine (CQ) and hydroxychloroquine (HCQ), have been utilized successfully in pre-clinical settings to mitigate tumorigenesis in various cancer types (4). Phase I/II clinical trials that examine the safety and therapeutic efficacy of CQ and HCQ in combination with various chemotherapies and targeted therapies are currently in progress (http://clinicaltrials.gov) (7,8). Dose-limiting toxicities of CQ and HCQ in certain patients, however, remain a clinical challenge (5,9). Other inhibitors that target core components of the autophagy machinery, like vacuolar protein sorting 34 (VPS34), unc-51 like autophagy activating kinase 1 (ULK1) and autophagy related 4B cysteine peptidase (ATG4B), have also been developed and used successfully in pre-clinical studies to mitigate tumorigenesis and to augment the effects of chemotherapies and targeted agents in various cancer models (4,5). To date, clinical trials examining their use in combination with chemotherapies and targeted agents have not yet been initiated.
Other recent efforts have turned to investigating the potential for targeting regulators of autophagy, particularly those that are upregulated or expressed specifically in cancer contexts. Emerging evidence has suggested that the eukaryotic elongation factor 2 kinase (eEF2K) promotes tumor survival and growth by negatively regulating protein synthesis when resources in the cell become limiting (10). The activity of eEF2K is tightly regulated by nutrient-sensing pathways, like mammalian target of rapamycin complex 1 (mTORC1) and adenosine monophosphate-activated protein kinase (AMPK). mTORC1 negatively regulates the activity of eEF2K by phosphorylating the kinase at distinct inhibitory sites (11,12). Of note, both AMPK and mTORC1 function in the regulation of autophagy (13). mTORC1 suppresses the autophagy pathway by phosphorylating and inhibiting ULK1, whereas AMPK promotes autophagy by inhibiting mTORC1 and phosphorylating and activating ULK1 (13). Under conditions of nutrient stress, the core cellular energy sensor AMPK phosphorylates and inhibits the activity of mTORC1 (11,14), and this consequently suppresses mTORC1-mediated eEF2K inhibition. Concurrently, AMPK phosphorylates and activates eEF2K, and this results in the subsequent phosphorylation and inhibition of the eEF2K downstream target, eukaryotic elongation factor 2 (eEF2) (11,15). The phosphorylation of eEF2 consequently mitigates protein synthesis by inhibiting eEF2-mediated translocation of nascent protein chains across the ribosome during elongation, and conserves cellular resources like amino acids and ATP (adenosine-5'-triphosphate) (10). In this manner, the role of eEF2K in energy conservation can be leveraged by cancers to meet their high energy and metabolic demands (13).
Depending on the type of cancer and/or stressor, eEF2K differentially regulates autophagy to promote survival under conditions of stress (10). For example, in cell line models of glioblastomas and breast cancers, eEF2K is required for the activation of pro-survival autophagy under conditions of endoplasmic reticulum (ER) stress (10,16). Cheng et al. demonstrated that the activation of eEF2K in response to ER stress is mediated by ER membrane-associated proteins involved in the unfolded protein response and the activating transcription factor 4 (ATF4) protein. Although ATF4 has been linked to the transcriptional upregulation of proteins that partake in autophagy, like microtubule associated protein 1 light chain 3 beta (MAP1LC3B, or LC3B), the molecular mechanisms underlying eEF2K-mediated autophagy and tumor cell survival in these cancer contexts remain undetermined (16). In contrast, eEF2K inhibition has also been associated with activation of pro-survival autophagy and reduced chemosensitivity to oxaliplatin in colon cancers (17). It has been proposed that inhibition of eEF2K de-represses protein synthesis, and this consequently activates AMPK and ULK1, and promotes autophagy and tumor cell survival (17). In other cancers like lung adenocarcinomas, eEF2K promotes tumorigenesis but does not appear to regulate autophagy. Moore et al. previously demonstrated that eEF2K inhibition elicits no effect on autophagic flux but impairs tumor cell survival under conditions of nutrient deprivation (11). When cellular glucose levels are scarce, eEF2K is activated to suppress and alleviate the high energy costs of protein synthesis, thereby facilitating the survival of lung adenocarcinoma cells (11). These studies present interesting observations regarding the context-dependent roles of eEF2K in autophagy regulation and cellular energy conservation in cancers.
A recent study in this journal by Wang et al. (18) has now linked eEF2K and autophagy to chemotherapy response in triple negative breast cancers (TNBC). To evaluate the role of autophagy in resistance to the chemotherapeutic agent paclitaxel, the authors first derived paclitaxel-resistant cell lines from two parental TNBC lines (MDA-MB-231 and MDA-MB-468) and both the parental and resistant cell lines were then treated with CQ. While both the parent and resistant lines were affected by CQ, the paclitaxel-resistant lines showed a more pronounced effect with respect to decreased viability, spheroid formation, and invasive potential (18). The combination of CQ with paclitaxel substantially reduced the IC50 values in the MDA-MB-231 and MDA-MB-468 paclitaxel-resistant lines by 12.6- and 3.9-fold, respectively, compared to 2.1 and 1.1 in the corresponding parental lines. These findings suggested that autophagy may play a role in modulating the sensitivity of TNBC cells to paclitaxel. As previous studies had suggested that eEF2K may be involved in the induction of autophagy (10,16), the authors next evaluated the role of eEF2K in regulating autophagy in paclitaxel-resistant TNBC cells. They showed that genetic knockdown of eEF2K reduced the levels of lipidated LC3B (LC3B-II) relative to control-shRNA treated cells under both fed and starvation conditions (18). As a reduction in LC3B-II protein levels may not necessarily be interpreted as a reduction in autophagic flux, the authors employed the use of a tandem fluorescent reporter mRFP-GFP-LC3B in an autophagy flux assay to further delineate if eEF2K knockdown induced or reduced autophagy levels. They showed that, in the presence of bafilomycin A1, eEF2K knockdown was associated with a reduction in levels of LC3B puncta (both red- and green-positive), indicating a reduction in autophagosome formation. This was consistent with the observed reduction in LC3B-II protein levels following eEF2K knockdown and, together, suggest that eEF2K is involved in promoting autophagy in paclitaxel-resistant TNBC cells (18). The authors also demonstrated that genetic inhibition of eEF2K sensitized resistant TNBC cell lines to paclitaxel, and was associated with a reduction in colony-forming ability and invasive potential. Together, these studies support a role for eEF2K in positively regulating autophagy, and modulating chemotherapy response and cell viability in paclitaxel-resistant TNBC cell lines. The mechanistic role, however, of eEF2K in the regulation of autophagy and its potential relationship to treatment response and cell viability in this context remains to be elucidated.
To examine the potential clinical relevance of eEF2K and LC3B expression, Wang et al. performed immunohistochemistry (IHC) analyses of 222 HER2-negative breast cancer specimens (18). The selected patient cohort consisted of individuals that underwent neoadjuvant chemotherapy regimens with paclitaxel and carboplatin but did not achieve pathologically complete responses (pCR). Of the 222 residual post-treatment patient tumors that were examined in this study, 65% were classified as luminal-like [HER2-negative and hormone receptor (HR)-positive], and 35% were classified as TNBC (HER2-negative and HR-negative). The histo-score (H-score) method was used to evaluate both LC3B and eEF2K staining. Positive expression of eEF2K was found to be more commonly associated with TNBC tumors compared to luminal-like tumors. In multivariate survival analyses, LC3B and eEF2K were each found to be independent predictors of disease-free survival (DFS). Patients with LC3B-positive or eEF2K-positive tumors had reduced DFS and these associations were more significant in the TNBC group. When analyzed in combination, the eEF2K-positive and LC3B-positive TNBC subgroup had the worst DFS, and patients with tumors that were negative for both eEF2K and LC3 had the best DFS (18).
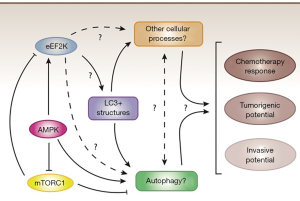
These findings together provide evidence supporting an association between eEF2K and LC3B in chemotherapy response in TNBC. To date, chemotherapeutic agents, like paclitaxel and other taxanes, remain part of first-line treatment regimens for patients diagnosed with TNBC (19). Chemotherapy resistance, however, is a frequent event, and patients often suffer from early recurrences and metastatic disease. The work by Wang et al. (18) identifies intriguing associations between eEF2K, LC3B, and treatment response that suggest eEF2K is a potential therapeutic target for paclitaxel-resistant TNBC. Future investigations are required to address important remaining questions (Figure 1) which include: (I) What are the molecular mechanisms underlying the role of eEF2K in autophagy regulation? Clarifying the molecular mechanisms underlying the role of eEF2K in autophagy regulation will help identify cancer contexts where eEF2K inhibition might suppress pro-survival autophagy and mitigate tumor progression; (II) Are the observed effects of genetic eEF2K inhibition in resistant TNBC cell lines a direct consequence of impairment in autophagic flux, or are other downstream processes involved? Alternatively, could the observed reduction in the formation of LC3B-positive structures from eEF2K inhibition be a consequence of impairment in autophagy-independent LC3B-related processes? Indeed, several studies have reported autophagy-independent roles of LC3 proteins that contribute to tumor progression (20), including packaging of extracellular vesicles (21,22) and anoikis (23); (III) Can the observed effect of pharmacological inhibition by CQ on paclitaxel response be recapitulated by genetic approaches that target key ATG genes? In other words, are the effects of CQ due to inhibition of autophagy or to inhibition of other lysosomal-related processes? (IV) Can the in vitro findings by Wang et al. be recapitulated in an in vivo setting and, more importantly, a pre-clinical setting? Several small molecule compounds that function as inhibitors of eEF2K have been developed and are currently being investigated in pre-clinical studies in various tumor models (24). It will be of value to test these inhibitors in murine models to potentially support their clinical translation given the described oncogenic roles of eEF2K in this study and others (10); (V) Although an association between positive eEF2K and LC3B expression and reduced DFS was identified in TNBC patients in this study, can they be independently validated in larger datasets to confirm their clinical relevance as negative prognostic biomarkers in TNBC patients? Will evaluation of LC3B puncta yield similar findings as the LC3B H-score? Will eEF2K and/or LC3B expression have utility as predictive biomarkers in TNBC patients? While many questions remain, their answers could help to identify and stratify TNBC patients that may potentially benefit from combination treatment strategies involving eEF2K inhibition and/or autophagy inhibition.
Acknowledgments
The authors thank Morgana Xu, Gayathri Samarasekera and Robert Camfield for helpful comments on the manuscript.
Funding: The authors gratefully acknowledge support from CIHR PJT-159536 grant to SMG. CJH is supported in part by a Simon Fraser University Molecular Biology and Biochemistry Graduate Fellowship.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2930). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Boumahdi S, de Sauvage FJ. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat Rev Drug Discov 2020;19:39-56. [Crossref] [PubMed]
- Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature 2019;575:299-309. [Crossref] [PubMed]
- Thorburn A, Thamm DH, Gustafson DL. Autophagy and Cancer Therapy. Mol Pharmacol 2014;85:830-8. [Crossref] [PubMed]
- Ho CJ, Gorski SM. Molecular Mechanisms Underlying Autophagy-Mediated Treatment Resistance in Cancer. Cancers 2019;11:1775. [Crossref] [PubMed]
- Amaravadi RK, Kimmelman AC, Debnath J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov 2019;9:1167-81. [Crossref] [PubMed]
- Towers CG, Wodetzki D, Thorburn A. Autophagy and cancer: Modulation of cell death pathways and cancer cell adaptations. J Cell Biol 2020;219:e201909033. [PubMed]
- Chude CI, Amaravadi RK. Targeting Autophagy in Cancer: Update on Clinical Trials and Novel Inhibitors. Int J Mol Sci 2017;18:1279. [Crossref] [PubMed]
- Zhan L, Li J, Wei B. Autophagy therapeutics: preclinical basis and initial clinical studies. Cancer Chemother Pharmacol 2018;82:923-34. [Crossref] [PubMed]
- Poklepovic A, Gewirtz DA. Outcome of early clinical trials of the combination of hydroxychloroquine with chemotherapy in cancer. Autophagy 2014;10:1478-80. [Crossref] [PubMed]
- Wang X, Xie J, Proud CG. Eukaryotic Elongation Factor 2 Kinase (eEF2K) in Cancer. Cancers (Basel) 2017;9:162. [Crossref] [PubMed]
- Moore CEJ, Wang X, Xie J, et al. Elongation factor 2 kinase promotes cell survival by inhibiting protein synthesis without inducing autophagy. Cell Signal 2016;28:284-93. [Crossref] [PubMed]
- Wang X, Regufe da Mota S, Liu R, et al. Eukaryotic Elongation Factor 2 Kinase Activity Is Controlled by Multiple Inputs from Oncogenic Signaling. Mol Cell Biol 2014;34:4088-103. [Crossref] [PubMed]
- Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132-41. [Crossref] [PubMed]
- Agarwal S, Bell CM, Rothbart SB, et al. AMP-activated Protein Kinase (AMPK) Control of mTORC1 Is p53- and TSC2-independent in Pemetrexed-treated Carcinoma Cells. J Biol Chem 2015;290:27473-86. [Crossref] [PubMed]
- Johanns M, Pyr Dit Ruys S, Houddane A, et al. Direct and indirect activation of eukaryotic elongation factor 2 kinase by AMP-activated protein kinase. Cell Signal 2017;36:212-21. [Crossref] [PubMed]
- Cheng Y, Ren X, Zhang Y, et al. Integrated regulation of autophagy and apoptosis by EEF2K controls cellular fate and modulates the efficacy of curcumin and velcade against tumor cells. Autophagy 2013;9:208-19. [Crossref] [PubMed]
- Xie CM, Liu XY, Sham KW, et al. Silencing of EEF2K (eukaryotic elongation factor-2 kinase) reveals AMPK-ULK1-dependent autophagy in colon cancer cells. Autophagy 2014;10:1495-508. [Crossref] [PubMed]
- Wang RX, Xu XE, Huang L, et al. eEF2 kinase mediated autophagy as a potential therapeutic target for paclitaxel-resistant triple-negative breast cancer. Ann Transl Med 2019;7:783. [Crossref] [PubMed]
- Nedeljković M, Damjanović A. Mechanisms of Chemotherapy Resistance in Triple-Negative Breast Cancer—How We Can Rise to the Challenge. Cells 2019;8:957. [Crossref] [PubMed]
- Galluzzi L, Green DR. Autophagy-Independent Functions of the Autophagy Machinery. Cell 2019;177:1682-99. [Crossref] [PubMed]
- Guo H, Chitiprolu M, Roncevic L, et al. Atg5 Disassociates the V1V0-ATPase to Promote Exosome Production and Tumor Metastasis Independent of Canonical Macroautophagy. Dev Cell 2017;43:716-30.e7. [Crossref] [PubMed]
- Leidal AM, Huang HH, Marsh T, et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat Cell Biol 2020;22:187-99. [Crossref] [PubMed]
- Satyavarapu EM, Das R, Mandal C, et al. Autophagy-independent induction of LC3B through oxidative stress reveals its non-canonical role in anoikis of ovarian cancer cells. Cell Death Dis 2018;9:934. [Crossref] [PubMed]
- Liu R, Proud CG. Eukaryotic elongation factor 2 kinase as a drug target in cancer, and in cardiovascular and neurodegenerative diseases. Acta Pharmacol Sin 2016;37:285-94. [Crossref] [PubMed]


