
Comparative proteomics analysis of plasma protein in patients with neuropsychiatric systemic lupus erythematosus
Introduction
Systemic lupus erythematosus (SLE) is a prototypic and complex inflammatory disorder featured by a variety of autoantibodies and multisystem involvement (1). Previous studies have shown that neurologic and neuropsychiatric (NP) symptoms occur in up to 75% of SLE patients, making NPSLE the most common and particularly severe form of lupus (2). The symptoms of NPSLE include mood disorders, psychosis, confusion, headache and cognitive dysfunction, which significantly degraded the quality of life and affect the survival of NPSLE patients (3,4). It was reported that NP events can adversely affect health related quality of life of SLE patients, regardless of disease activity, organ involvement and drug exposure (5). However, the mechanisms underlying NPSLE pathogenesis remain limited, so no single test combining high specificity with high sensitivity is sufficient for diagnosis of NPSLE (2). Therefore, there is an urgent need to evaluate new biomarkers of NPSLE for early diagnosis, prognosis and treatment.
As a source of new biomarkers for lots of diseases, proteomics technology has increasingly maintained attention over the past two decades (6,7). To date, tandem mass tags (TMT) based quantitative proteomics technology has been widely applied in the protein biomarker research and protein alterations quantification in many autoimmune diseases and cancers (8,9). Among the previous proteomic studies about autoimmune diseases, SLE has received some degree of attention (10-12). However, no TMT technology has been reported involved in plasma related to patients with SLE or NPSLE.
Therefore, the current study quantitatively analyzed the proteomics profiling of plasma in NPSLE, active SLE without central nervous system involvement (non-NPSLE) and healthy controls (HC). The goal of this research was to discover the protein changes possibly related with the disease progression, or a biomarker to prompt diagnosis or predict prognosis of NPSLE. So, a TMT-liquid chromatography-triple quadrupole mass spectrometry (LC–MS/MS)-based proteomics method was used to define the differential plasma proteome across plasma samples from NPSLE patients to investigate possible molecular mechanisms involved in NPSLE and pave the way for the novel potential therapeutic targets.
Methods
Patients and healthy controls
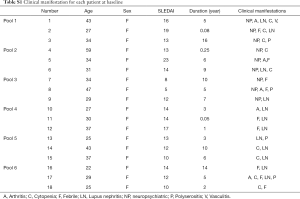
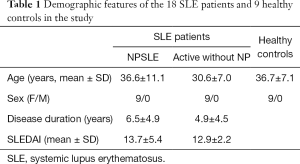
In total, eighteen patients with SLE and nine healthy controls were invited to participate in our research. Lupus patients were divided into two groups, 9 patients with NPSLE and 9 non-NPSLE patients. Detailed information of these patient samples is shown in the Table S1. All patients were diagnosed according to the American College of Rheumatology (ACR) revised SLE criteria (13). Disease activity of these patients was measured using the Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) (14). NPSLE patients were fulfilled with the ACR nomenclature and case definitions for NPSLE (15).
Full table
Altogether 27 plasma samples were collected. 2 mL of whole blood was collected separately from study subjects and K2-EDTA was used as an anticoagulant. All samples were then centrifuged at room temperature at 1,300 ×g for 10 min. The resulting plasma was collected and then kept at −80 °C before used. Within each group, every three plasmas were mixed into single samples in order to obtain three pooled samples.
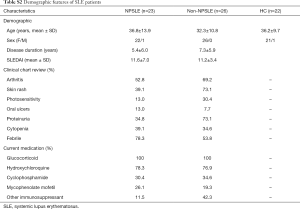
Validation was performed using a second larger and independent sample set. Totally 23 NPSLE, 26 non-NPSLE and 22 age-matched healthy controls were included. Demographic features of these patient samples are shown in the Table S2. The sample cohorts were matched for age and gender. This research was approved by the Ethics Committee at The Affiliated Drum Tower Hospital of Nanjing University Medical School (ID: SC201700201) and was undertaken according to the guidelines of the Declaration of Helsinki. Written informed consent was obtained from the patient for publication of this study and any accompanying images.
Full table
Plasma sample preparation and SDS-PAGE
High-abundance proteins was eliminated using the Agilent Multiple Affinity Removal System (Agilent, USA) in this study, according to the manufacturer’s suggestions. The depleted plasma samples were washed three times with 0.5 mL of 25 mM Ammonium bicarbonate, concentrated to 100 µL and rapidly transferred to 1.5 mL polypropylene centrifuge tubes. Each tube was added with 0.1 mL SDT protein lysis buffer and then heated with 100 °C metal bath for 3 min, following by cooling with ultrasonic in an ice bath (50 W for 2 s, interval 8 s, total sonication time 5 min). The samples were heated with 100 °C metal bath for 3 min again. The final protein concentration was measured using a Bradford assay kit (BioRad, Hercules, CA). Subsequently, samples (20 µg) were taken for SDS-PAGE according to the quantitative data. Depletion of each set of plasma samples was completed in the same day so that all the samples were processed consistently in the same set. The protein extracts were stored at –80 °C.
Protein digestion
The protein samples were digested in standard sample buffer by FASP procedure (16). The proteins mixture was reduced for 5 min at 100 °C with 100 mM DTT. After cooling to room temperature, 200 µL of UA buffer (pH 8.5) containing 150 mM Tris-HCl and 8M Urea was added and mixed well. All samples were transferred to ultrafiltration with a 30 kDa cut-off membrane filter (Sartorius, Gottingen, Germany) and centrifuged for 30 min at 14,000 ×g, and then discarded the filtrate. This step was repeated for 3 times. Subsequently, the proteins were alkylated with 100 µL IAA (50mM IAA in UA), shaked for 1 min at 600 rpm and incubated in the dark for 30 min at 300 rpm. Then, each tube was centrifuged for 30 min at 14,000 ×g. Add 100 µL UA buffer, centrifuge for 30 min at 14,000 ×g. This step was repeated for 3 times. 100 µL dissolution buffer (100 mM/L) (Applied Biosystems, USA) was added, and centrifuged for 30 min at 14,000 ×g at room temperature (RT). This step was repeated for 3 times. Finally, the filtrate was discarded and the proteins were digested with trypsin on a thermostat mixer at 300 rpm for 18 h at 37 °C.The tryptic peptides were centrifuged (14,000 ×g, 30 min, RT), replaced with new tube, and then added with 40 µL of 25 mM DS buffer. The tubes were centrifuged (14,000 ×g, 30 min, RT) and the filtrate was collected. Finally, the tryptic peptides were quantified by OD280 (16).
TMT Labeling and high-pH reversed-phased chromatography separation
Three pooled NPSLE patient sera samples, 3 pooled non-NPSLE patient sera samples and 3 pooled healthy control sera samples, with each pool comprised of sera aliquots from 3 individuals (total of 9 healthy control and 18 SLE patients), were included in this global proteomics discovery study. Digested peptides were labeled with TMT reagents (Thermo Scientific) following procedures recommended by the manufacturer. All labeled peptides were pooled together. 1,100 Series HPLC Value System (Agilent) equipped with a Gemini-NX (Phenomemex, 00F-4453-E0) column (4.6 × 150 mm, 3 µm, 110 Å) was used for high-pH reverse-phase HPLC to fractionate peptide samples. Peptides were separated into 40 fractions using a gradient of 4.5% to 90% ACN in 10mMammonium bicarbonate (pH 10) over 130 min. The peptides were then combined into 18 fractions and dried by vacuum centrifugation. Samples were storaged at −80 °C freezer for further analysis.
LC-MS/MS analysis
Easy-nLC nanoflow HPLC system connected to Orbitrap-Fusion (Thermo Fisher Scientific, San Jose, CA, USA) were used to analyze the TMT-labeled samples. Peptides were reconstituted in 0.1% v/v trifluoroacetic acid. Each sample (1 µg) was loaded onto Thermo Scientific EASY column (two columns) using an autosampler at a flow rate of 200 nL/min. Peptides were accomplished using a segmented gradient from 5% to 28% Solvent B (0.1% formic acid in 100% ACN) for 40 min, followed by 28–90% Solvent B for 2 min and then 90% Solvent B for 18 min. The column was re-equilibrated to its initial highly aqueous solvent composition before each analysis. LC-MS/MS analysis was performed on an Obitrap Fusion mass spectrometer (Thermo Scientific). The MS scan and MS/MS scan were acquired at a resolution of 1.2×105 and 5×104 at 200 m/z, respectively. The maximum ion injection times were maintained at 105 ms for the MS/MS scans, and automatic gain control target values for Master scan modes was set to 4×105 and 1×105.
Database search and protein quantification
Using the Proteome Discoverer 2.1 software, acquired MS/MS spectra were analyzed for protein identification and quantification. Searching for the fragmentation spectra was carried out by the MASCOT search engine embedded in Proteome Discoverer against the Uniprot human database (March 8, 2017, 156914 sequences). A false discovery rate (FDR) of no more than 1% was used for data filtering.
Using Proteome Discoverer (version 2.1), the analysis of the proteins based on the ratios of TMT reporter ions from all unique peptides representing each protein was performed using Proteome Discoverer (version 2.1) in the samples. The peak intensities of the TMT reporter ions released in each of the MS/MS spectra were used, and the IS sample was used as a reference in analyzing for the TMT ratios of the reporter ions. Finally, the median of the unique peptides of the protein by calculating relative protein quantifications were first normalized.
Bioinformatics analysis
The Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) database were applied to explore the potential roles of differentially expressed proteomics by DAVID 6.8 (https://david.ncifcrf.gov/). The proteins were classified for the next three categories: molecular function, cellular component, and biological process.
ELISA experiments
Validation of differentially expressed proteomics results was performed using independent sample from the same cohort in proteomic analysis. Comprehensive considering proteomics profiles and function of proteins, we chose to verify four proteins measured by ELISA, respectively, including C1q/TNF-related protein-3 (CTRP3), vitamin D binding protein (VDBP), pregnancy-associated plasma protein-A (PAPPA) and serine protease 2 (TRYP2). Serum concentrations of CTRP3, VDBP, PAPPA and TRYP2 were all measured using commercially available ELISA kits. CTRP3 and VDBP ELISA kits were from R&D Systems. PAPPA and TRYP2 ELISA kits were from RayBiotech. We detected the serum concentrations according to the manufacturer’s instructions.
Statistics
All data were presented as mean ± SD. Differentially expressed proteins were identified by the fold of change (fold >1.5) method and independent t-test. Differences between two groups were determined by unpaired Student’s t-test if the variance was normally distributed. Comparisons among three or more groups were conducted using one-way ANOVA. Randomization and blinding strategy was used whenever possible. The correlations were analyzed by Spearman’s correlation test. Data were analyzed and visualized with SPSS16.0 software or GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, USA) and a two-tailed P value <0.05 was considered statistically significant.
Results
Protein profile changes in NPSLE patients identified by MS/MS analysis
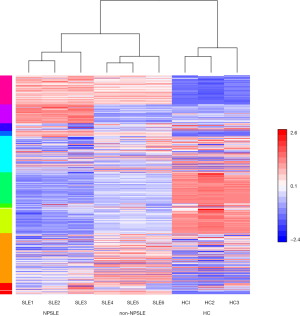
To determine the possible role of protein profiles in NPSLE pathogenesis, we performed a TMT-labeled quantitative proteomic study to compare the protein profile differences among NPSLE patients, non-NPSLE patients and healthy subjects. Information for the samples is shown in Table 1. A total of 726 proteins were identified in our proteomic studies between SLE and healthy controls using scatter plot and hierarchical clustering analyses (Figure 1). The original raw data were showed in the in total online: http://fp.amegroups.cn/cms/aced7f51ea115b5ba81dd6fdf54db336/atm.2020.04.58-1.xlsx. Hierarchical clustering demonstrated that two groups were clustered based on the protein abundance levels of the samples: proteins of NPSLE and non-NPSLE clustered together, whereas those of HC clustered independently.
Full table
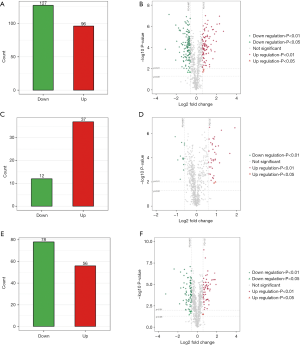
Further, the greatest number of proteins with different abundances occurred when analyzing NPSLE and HC samples. There were fewest proteins with significantly different abundances in NPSLE/non-NPSLE comparisons. More specifically, compared with HC group, 96 proteins were increased and 127 were decreased (cutoff ratio of >1.50 and <0.667) among proteins identified (Figure 2A) in the NPSLE group. The details of proteins differently expressed between NPSLE and non-NPSLE groups were showed in the in total online: http://fp.amegroups.cn/cms/d75d5c8a7207877825537895b750a77a/atm.2020.04.58-2.xlsx. The volcano plot (Figure 2B) depicted the distribution of proteins according to statistical significance (P values) and NPSLE/HC abundance ratio. Compared with non-NPSLE group, 37 proteins were increased and 12 were decreased (cutoff ratio of >1.50 and <0.667) in NPSLE/ non-NPSLE group (Figure 2C), so did the volcano plot show (Figure 2D). The details of proteins differently expressed between NPSLE and HC groups were showed in the in total online: http://fp.amegroups.cn/cms/f965d561360731ff332d3f13d6566ecc/atm.2020.04.58-3.xlsx. Similarly, 56 proteins were increased and 78 were decreased (cutoff ratio of >1.50 and <0.667) in non-NPSLE group compared to that of HC group (Figure 2E), so did the volcano plot show (Figure 2F). The details of proteins differently expressed between non-NPSLE and HC groups were showed in the in total online: http://fp.amegroups.cn/cms/6acf489810c38ddc194d3a3802d58dd9/atm.2020.04.58-4.xlsx.
Verification of differentially expressed proteins by ELISA
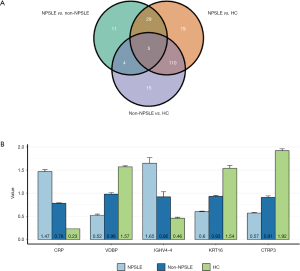
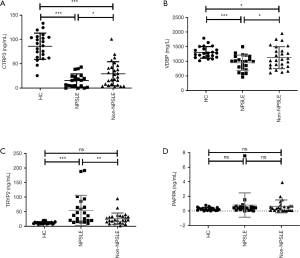
To verify the differentially expressed proteins which were identified by TMT-labeled LC-MS/MS analysis, four aberrantly expressed proteins were selected for further analysis by ELISA in the serums from NPSLE patients, non-NPSLE patients and healthy subjects. First, we analysis the differentially expressed proteins between two different groups by Venn analysis, indicating that only five proteins were got using the following criteria: tendencies of abnormally expressed proteins were consistent between NPSLE-non-NPSLE group and between non-NPSLE-HC group (Figure 3). The five proteins were showed in Figure 4, including VDBP, C-reactive protein (CRP), Keratin 16 (KRT16), Immunoglobulin heavy variable 4-4 (IGHV4-4) and CTRP3. The annotative MS spectra for the unique peptides of 5 identified proteins were provided in the Figure S1. In view of not all proteins have known function, we chose VDBP and CTRP3 to verify. In addition, to further analyze the difference between NPSLE and non-NPSLE, we selected two proteins (PAPPA and TRYP2) that significantly changed between NPSLE/non-NPSLE and NPSLE/HC comparisons, but with no significant difference between non-NPSLE/HC comparisons. As can be seen from Figure 4A, the expression of CTRP3 levels was significantly decreased in patients with NPSLE compared with patients with non-NPSLE (P<0.05) and healthy controls (P<0.001). CTRP3 levels were also significantly decreased in patients with non-NPSLE compared with healthy controls (P<0.001). The similar results could be seen in the expression of VDBP levels in three groups (Figure 4B). The expression of the TRYP2 levels was significantly increased in patients with NPSLE compared with patients with non-NPSLE (P<0.01, Figure 4C) and healthy controls (P<0.001, Figure 4C). There was no difference between patients with non-NPSLE and healthy controls (P>0.05, Figure 4C). PAPPA levels were not dysregulated between these groups (P>0.05, Figure 4D).
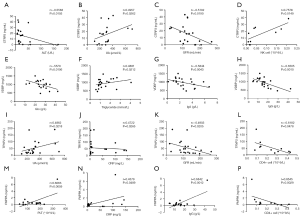
Next, to define potential clinical effects of those proteins in NPSLE, we analyzed the correlations between the differentially expressed proteins levels and the clinical characteristics of NPSLE patients. As showed in Figure 5A,B,C,D, the expression levels of CTRP3 were associated with aminotransferase (ALT), uric acid (UA), glomerular filtration rate (GFR) and NK cells numbers (P<0.05). Whereas, VDBP levels were found to be associated with globin (Glo), triglyceride, serum IgA and IgG levels (P<0.05, Figure 5E,F,G,H). Furthermore, the expression levels of TRYP2 were associated with UA, CRP, GFR and CD4+ cells numbers (P<0.05, Figure 5I,J,K,L). The expression levels of PAPPA were associated with platelets (PLT), CRP, serum IgG levels and CD4+ cells numbers (P<0.05, Figure 5M,N,O,P).
Gene ontology (GO) analysis
The GO data were used to examine the subcellular annotation of totally expressed proteins and the function of the differentially expressed proteins.
Subcellular location annotation of totally proteins
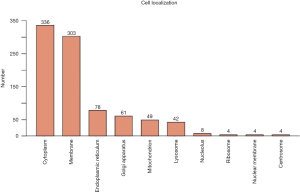
As showed in Figure 6, the totally expressed proteins were quantified according to the subcellular location annotation. Most proteins were concentrated in the following categories: cytoplasm, membrane, endoplasmic reticulum (37.8%, 34.1% and 8.8%, respectively).
Evaluation of differentially expressed proteins by GO analysis
To further evaluate the potential biological significance of the 726 proteins identified in three groups, GO enrichment analysis was employed to analyze the proteins. As shown in Figure 7, the top 10 GO items from the differentially expressed proteins between the different groups were listed. Our data demonstrated that the most significantly enriched cellular components of the differentially expressed proteins in serum of NPSLE and non-NPSLE were extracellular region, extracellular region part and membrane-bounded vesicle (Figure 7A). The most significantly enriched cellular components of NPSLE and healthy controls were extracellular space, extracellular region part and extracellular exosome (Figure 7B). The most significantly enriched cellular components of non-NPSLE and healthy controls were extracellular vesicle, extracellular organelle and extracellular region part (Figure 7C).
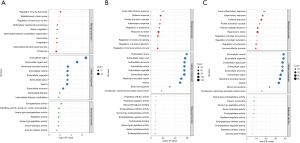
As for biological processes, the most significantly enriched item of NPSLE and non-NPSLE were regulation of body fluid levels, establishment of skin barrier and regulation of water loss via skin (Figure 7A). The most significantly enriched biological processes of NPSLE and healthy controls were acute inflammatory response, defense response and protein activation cascade (Figure 7B). The most significantly enriched biological processes of non-NPSLE and healthy controls were acute inflammatory response, inflammatory response and defense response (Figure 7C).
As for molecular functions, the most significantly enriched item of NPSLE and non-NPSLE were endopeptidase activity, peptidase activity and serine type endopeptidase activity (Figure 7A). The most significantly enriched molecular functions of NPSLE and healthy controls were peptidase inhibitor activity, peptidase regulator activity and glycosaminoglycan binding (Figure 7B). The most significantly enriched molecular functions of non-NPSLE and healthy controls were serine-type endopeptidase activity and calcium ion binding and serine-type peptidase activity (Figure 7C). These together reveal the roles of significantly dysregulated proteins in NPSLE.
KEGG pathway enrichment analysis
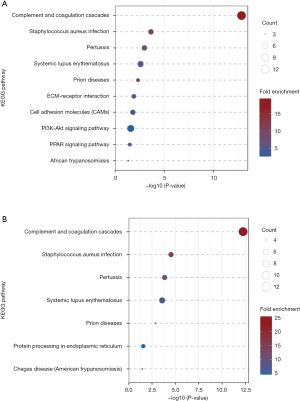
The KEGG pathway enrichment analysis was performed to identify the statistically significant differently expressed proteins (Figure 8) (P<0.05). Proteins in NPSLE and controls were mainly involved in complement and coagulation cascades. The same finding was in non-NPSLE and controls. There was no significantly changed pathway in NPSLE and non-NPSLE.
Discussion
Proteins are the direct function executors of myriad life activities (17). Compared to genomics, proteomics is highly dynamic and regulated by multiple factors, including protein expression/degradation, subcellular localization, post-translational modifications (PTMs) and proteins-proteins interactions and other biomolecules (18). Therefore, fast, high sensitivity and high throughput proteomics technology is of great significance. In recent years, proteomics technology combined with mass spectrometry has achieved extraordinary improvements and been widely used in different clinically orientated studies (7). Chemical labeling with isobaric TMT, including TMT and iTRAQ, has been the most popular methods among the quantitative proteomic technology (19,20). Wang et al. established protein expression profiles of PBMC from patients with SLE and healthy subjects with iTRAQ-based technology and identified 67 unique proteins (21). Similarly, Wang et al. also found that STRAP, an important inhibitor of TGF-ß signaling, decrease in active SLE patients compared with healthy controls using iTRAQ-MS (11). In another study, renal tissue from patients with lupus nephritis and healthy subjects was analyzed by iTRAQ-MS and four proteins were detected, including up-regulated hnRNP-, Annexins and down-regulated ASS, aldolase (12). Therefore, we identified differently expressed proteins using TMT-labeled LC-MS/MS analysis in serum from NPSLE, non-NPSLE patients and healthy controls in this study.
Neurological and psychiatric features are a serious complication of SLE, which can result in severe neurodegenerative changes and threaten life (22). It is reported that infection, lupus nephritis, hematological abnormality, NPSLE, and interstitial pneumonia are top causes of deaths in China (23). The two recognized underlying mechanisms of the immunopathogenesis of NPSLE were ischemic and inflammatory mechanisms, including blood-brain barrier dysfunction and production of pro-inflammatory cytokines which mediated by autoantibody (5,24). However, NPSLE pathogenesis is incompletely understood. It was reported that many autoantibodies have been linked to NPSLE, including anti-NR2 and anti-ribosomal P antibodies. However, there is no enough evidence to confirm involvement in mechanisms of pathogenesis (25). S100 β were increased in serum of adults and children with NPSLE, suggesting S100 β as a biomarker (26). Nevertheless, these biomarkers lack disease specificity of NPSLE. Nevertheless, these biomarkers lack disease specificity of NPSLE. There is a desperate need for discovering reliable NPSLE biomarkers that can be used for diagnosis, disease monitoring and better prediction of response to therapy. Therefore, we investigated the abnormally expressed proteins between NPSLE patients and non-NPSLE patients. Totally 726 proteins involving different cellular localizations and biological functions were identified in our proteomic studies. Further analysis, we found higher numbers of differentially expressed proteins in NPSLE/HC comparisons than those in non-NPSLE/HC comparisons, indicating that the inflammatory damage might be progressive in NPSLE.
Then, we found only five proteins significantly changed in all three groups-comparisons by Venn analysis, including VDBP, CRP, KRT16, IGHV4-4 and CTRP3. CRP, a biomarker of inflammation, was increased in patients with SLE both in remission and active periods when compared with control subjects (27). However, the levels of CRP were not always related to SLE disease activity. The significance of CRP remains controversial and is not one of the recommended tools in SLE disease activity follow-up (28).The study on KRT16 and IGHV4-4 concerning lupus is rare. Above all, we did not verify the three proteins. VDBP (also known as Gc-globulin) is a multifunctional protein and the major plasma carrier protein of vitamin D and its metabolites (29). In our study, we found that serum VDBP levels were significantly decreased in SLE patients compared with healthy controls, and they were lower in patients with NPSLE than with non-NPSLE. It was reported that vitamin D deficiency may contribute towards the pathogenesis of SLE (30). Our result, along with other reported findings, suggests decreased levels of vitamin D, decreased levels of VDBP, or both may participate in the pathogenesis and process of SLE. However, studies on VDBP in NPSLE were rare. Our results showed that the levels of VDBP in NPSLE were significantly decreased compared with non-NPSLE, which indicate that VDBP may participate in the pathogenesis of NPSLE. Nevertheless, the detailed role it plays in NPSLE still needs further study.
CTRP3, a member of C1q TNF Related Protein (CTRP) family, plays important roles in modulating glucose and lipid metabolism (31). Recent studies have demonstrated that CTRP3 has anti-inflammatory effects in attenuating LPS-induced systemic inflammation (32). Moreover, the deficiency of CTRP3 markedly exacerbated inflammation in a mouse model of rheumatoid arthritis (33). In our study, we found that the expression of CTRP3 were extremely decreased in patients with NPSLE when compared with patients without NPSLE and healthy subjects. As cardiovascular disease is one of the leading causes of morbidity and mortality in patients with SLE and dyslipidemia in systemic lupus erythematosus has gained extensive attention (34), the role of CTRP3 that plays in SLE is worth further studying.
In addition, we also selected two proteins, PAPPA and TRYP2, which significantly changed between NPSLE/non-NPSLE and NPSLE/HC comparisons, but with no significant difference between non-NPSLE/HC comparisons. PAPPA, a member of the metzinc in metalloproteinase superfamily, was reported that exerts a proatherogenic effect in atherosclerosis (35). Besides, it was report that PAPPA was associated with the outcome of ischemic cerebrovascular disease (36). However, we only found that there was a tendency to increase of PAPPA in NPSLE. What’s more, we also detected the expression of PAPPA in cerebrospinal fluid in SLE patients and the results were consistent with in the serums. As we know, one underlying mechanisms of NPSLE progression was ischemic. We believed that more studies should be conducted to explore the mechanism by which PAPPA might be involved in the pathogenesis of NPSLE. There are few studies on trypsin 2 (TRYP2) in NPSLE, mainly focusing on tumors, and more research is needed to discover the role of TRYP2 in NPSLE.
In KEGG analysis, the pathway of complement and coagulation cascades has been emphasized between not only NPSLE-HC but also non-NPSLE-HC. The role of the complement pathway in lupus has been widely reported, and our research has shown that the complement pathway plays a critically important role not only in non-NPSLE patients, but also in NPSLE patients (37-39). Unfortunately, there was no significantly changed pathway in NPSLE and non-NPSLE in our study.
In conclusion, our study has successfully increased serum protein profiles of NPSLE, non-NPSLE patients and healthy controls through TMT technology and screened out five proteins significantly changed in NP-NPSLE patients. The pathway of complement and coagulation cascades may participate in pathogenesis of NPSLE.
Acknowledgments
Funding: The work was supported by National Natural Science Foundation of China (No. 81571586, 81871283 and 81801555), Research Project of Jiangsu Province Health Committee (H2019060), Medical Science and Technology Development Key Project of Nanjing (ZKX15018), and Jiangsu Province Six Talent Peaks Project (No. 2016-WSN-148).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.04.58). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by Institutional Ethics Committee of the Affiliated Drum Tower Hospital of Nanjing University Medical School and followed the tenets of the Declaration of Helsinki.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tipton CM, Fucile CF, Darce J, et al. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol 2015;16:755-65. [Crossref] [PubMed]
- Jeltsch-David H, Muller S. Neuropsychiatric systemic lupus erythematosus: pathogenesis and biomarkers. Nat Rev Neurol 2014;10:579-96. [Crossref] [PubMed]
- Wen J, Stock AD, Chalmers SA, et al. The role of B cells and autoantibodies in neuropsychiatric lupus. Autoimmun Rev 2016;15:890-5. [Crossref] [PubMed]
- Hanly JG, Urowitz MB, Su L, et al. Prospective analysis of neuropsychiatric events in an international disease inception cohort of patients with systemic lupus erythematosus. Ann Rheum Dis 2010;69:529-35. [Crossref] [PubMed]
- Hanly JG, Kozora E, Beyea SD, et al. Review: Nervous System Disease in Systemic Lupus Erythematosus: Current Status and Future Directions. Arthritis Rheumatol 2019;71:33-42. [Crossref] [PubMed]
- Zhang L, Elias JE. Relative Protein Quantification Using Tandem Mass Tag Mass Spectrometry. Methods Mol Biol 2017;1550:185-98. [Crossref] [PubMed]
- Moulder R, Bhosale SD, Goodlett DR, et al. Analysis of the plasma proteome using iTRAQ and TMT-based Isobaric labeling. Mass Spectrom Rev 2018;37:583-606. [Crossref] [PubMed]
- Cheng Y, Chen Y, Sun X, et al. Identification of potential serum biomarkers for rheumatoid arthritis by high-resolution quantitative proteomic analysis. Inflammation 2014;37:1459-67. [Crossref] [PubMed]
- Li GC, Zhang L, Yu M, et al. Identification of novel biomarker and therapeutic target candidates for acute intracerebral hemorrhage by quantitative plasma proteomics. Clin Proteomics 2017;14:14. [Crossref] [PubMed]
- Zhong S, Shu S, Wang Z, et al. Enhanced homing of mesenchymal stem cells to the ischemic myocardium by ultrasound-targeted microbubble destruction. Ultrasonics 2012;52:281-6. [Crossref] [PubMed]
- Wang L, Dai Y, Peng W, et al. Differential expression of serine-threonine kinase receptor-associated protein in patients with systemic lupus erythematosus. Lupus 2011;20:921-7. [Crossref] [PubMed]
- Sui W, Tang D, Zou G, et al. Differential proteomic analysis of renal tissue in lupus nephritis using iTRAQ reagent technology. Rheumatol Int 2012;32:3537-43. [Crossref] [PubMed]
- Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997;40:1725. [Crossref] [PubMed]
- Bombardier C, Gladman DD, Urowitz MB, et al. Derivation of the SLEDAI. A disease activity index for lupus patients. Arthritis Rheum 1992;35:630-40. [Crossref] [PubMed]
- The American College of Rheumatology nomenclature and case definitions for neuropsychiatric lupus syndromes. Arthritis Rheum 1999;42:599-608. [Crossref] [PubMed]
- Wiśniewski JR, Zougman A, Nagaraj N, et al. Universal sample preparation method for proteome analysis. Nat Methods 2009;6:359-62. [Crossref] [PubMed]
- Bai Y, Luo Q, Liu J. Protein self-assembly via supramolecular strategies. Chem Soc Rev 2016;45:2756-67. [Crossref] [PubMed]
- Leitner A. A review of the role of chemical modification methods in contemporary mass spectrometry-based proteomics research. Anal Chim Acta 2018;1000:2-19. [Crossref] [PubMed]
- Evans C, Noirel J, Ow SY, et al. An insight into iTRAQ: where do we stand now? Anal Bioanal Chem 2012;404:1011-27. [Crossref] [PubMed]
- Rauniyar N, Yates JR 3rd. Isobaric labeling-based relative quantification in shotgun proteomics. J Proteome Res 2014;13:5293-309. [Crossref] [PubMed]
- Wang L, Dai Y, Qi S, et al. Comparative proteome analysis of peripheral blood mononuclear cells in systemic lupus erythematosus with iTRAQ quantitative proteomics. Rheumatology international 2012;32:585-93. [Crossref] [PubMed]
- Dias-Santos A, Proenca RP, Tavares Ferreira J, et al. The role of ophthalmic imaging in central nervous system degeneration in systemic lupus erythematosus. Autoimmun Rev 2018;17:617-24. [Crossref] [PubMed]
- Wu XY, Yang M, Xie YS, et al. Causes of death in hospitalized patients with systemic lupus erythematosus: a 10-year multicenter nationwide Chinese cohort. Clin Rheumatol 2019;38:107-15. [Crossref] [PubMed]
- Magro-Checa C, Beaart-van de Voorde LJ, Middelkoop HA, et al. Outcomes of neuropsychiatric events in systemic lupus erythematosus based on clinical phenotypes; prospective data from the Leiden NP SLE cohort. Lupus 2017;26:543-51. [Crossref] [PubMed]
- Moore E, Huang MW, Putterman C. Advances in the diagnosis, pathogenesis and treatment of neuropsychiatric systemic lupus erythematosus. Current opinion in rheumatology 2020;32:152-8. [Crossref] [PubMed]
- Noris-García E, Arce S, Nardin P, et al. Peripheral levels of brain-derived neurotrophic factor and S100B in neuropsychiatric systemic lupus erythematous. Lupus 2018;27:2041-9. [Crossref] [PubMed]
- Dima A, Opris D, Jurcut C, et al. Is there still a place for erythrocyte sedimentation rate and C-reactive protein in systemic lupus erythematosus? Lupus 2016;25:1173-9. [Crossref] [PubMed]
- Mosca M, Tani C, Aringer M, et al. European League Against Rheumatism recommendations for monitoring patients with systemic lupus erythematosus in clinical practice and in observational studies. Ann Rheum Dis 2010;69:1269-74. [Crossref] [PubMed]
- Karras SN, Koufakis T, Fakhoury H, et al. Deconvoluting the Biological Roles of Vitamin D-Binding Protein During Pregnancy: A Both Clinical and Theoretical Challenge. Front Endocrinol (Lausanne) 2018;9:259. [Crossref] [PubMed]
- Nguyen MH, Bryant K, O'Neill SG. Vitamin D in SLE: a role in pathogenesis and fatigue? A review of the literature. Lupus 2018;27:2003-11. [Crossref] [PubMed]
- Wolf RM, Steele KE, Peterson LA, et al. Lower Circulating C1q/TNF-Related Protein-3 (CTRP3) Levels Are Associated with Obesity: A Cross-Sectional Study. PLoS One 2015;10:e0133955. [Crossref] [PubMed]
- Kopp A, Bala M, Buechler C, et al. C1q/TNF-related protein-3 represents a novel and endogenous lipopolysaccharide antagonist of the adipose tissue. Endocrinology 2010;151:5267-78. [Crossref] [PubMed]
- Petersen PS, Wolf RM, Lei X, et al. Immunomodulatory roles of CTRP3 in endotoxemia and metabolic stress. Physiol Rep 2016. [Crossref] [PubMed]
- Szabó MZ, Szodoray P, Kiss E. Dyslipidemia in systemic lupus erythematosus. Immunol Res 2017;65:543-50. [Crossref] [PubMed]
- Yu XH, He LH, Gao JH, et al. Pregnancy-associated plasma protein-A in atherosclerosis: Molecular marker, mechanistic insight, and therapeutic target. Atherosclerosis 2018;278:250-8. [Crossref] [PubMed]
- Wang S, Jiang J, Qu C, et al. Predictive value of serum pregnancy-associated plasma protein A for patients with ischemic cerebrovascular disease. J Clin Lab Anal 2017. [Crossref] [PubMed]
- Boackle SA. The Role of Complement in SLE. In: Wallace D, Hahn B. Dubois' Lupus Erythematosus and Related Syndromes. Elsevier, 2019:224-36.
- Magro-Checa C, Schaarenburg R, Beaart H, et al. Complement levels and anti-C1q autoantibodies in patients with neuropsychiatric systemic lupus erythematosus. Lupus 2016;25:878-88. [Crossref] [PubMed]
- Grossman TR, Hettrick LA, Johnson RB, et al. Inhibition of the alternative complement pathway by antisense oligonucleotides targeting complement factor B improves lupus nephritis in mice. Immunobiology 2016;221:701-8. [Crossref] [PubMed]


