
Differentiate cavernous hemangioma from schwannoma with artificial intelligence (AI)
Introduction
Cavernous hemangioma is one of the most common primary tumors that occur in the orbit, accounting for 3% of all orbital lesions (1-3), while schwannoma is a benign orbital tumor with a prevalence of less than 1% among all orbital lesions (1). It is necessary to distinguish these two tumors at treatment onset because they have different treatment strategies (4-6): complete removal is the treatment goal for cavernous hemangioma while for schwannoma, the goal is to ensure that no capsules remain. Moreover, clear differentiation provides useful information that fosters better vessel management (2). If the wrong surgical regimen is chosen, the tumor will recur, and the patient would need to undergo an additional operation.
Similar to the diagnosis of many other tumors, imaging techniques are applied as the predominant methods to diagnose these two tumors. Magnetic resonance imaging (MRI) is the most commonly used approach because of its high resolution, which clearly reflects the tissues to determine the appropriate surgical approach (5-7). However, because it manifests similarly to cavernous hemangioma, especially in MRI images, schwannoma often evokes an improper diagnosis (2,7,8) even highly experienced ophthalmologists or radiologists can make inaccurate diagnoses (9).
In recent years, the application of artificial intelligence (AI) in medicine has achieved physician-equivalent classification accuracy in the diagnosis of many diseases, including diabetic retinopathy (10-13), lung diseases (14), cardiovascular disease (15), liver disease, skin cancer (16), and thyroid cancer (17) and others.
Therefore, the goal of this project was to develop an AI framework that uses MRI image sets from 45 hospitals in China as input to automate the differentia diagnosis between cavernous hemangioma and schwannoma with high accuracy, sensitivity and specificity.
Methods
Overall architecture
Considering the current dominance of MRI in the differential diagnosis of the two studied tumor types, we selected MRI images as the research materials in this study. The research framework included of three types of functional models. Each type consists of eight groups of models with different arrangements and combinations of slice orientations (coronal and transverse) as well as weighted sequences (T1-weighted, T1-weighted contrast-enhanced, T2-weighted and T2-weighted fat suppression). Each group was sorted into 4 models and trained according to the principle of four-fold cross validation. In summary, a total of 96 models were obtained (3×8×4=96) (Figure 1).
As mentioned above, we established 3 types of functional models to achieve the goal of distinguishing cavernous hemangioma from schwannoma. First, to reduce interference from unnecessary information, eye-positioning models were designed to identify the eye area from the complete images. Then, to further narrow the recognition range, tumor-positioning models were created to locate tumors within the identified eye area. Finally, tumor classification models were trained to classify the tumors. As shown in Figure 2, when an MRI image is input, the framework first delineates the eye area from the whole image; then it localizes the tumor scope from the eye area; and finally, it specifically classifies the tumor. The eye-positioning and tumor-positioning models were trained using the Faster-RCNN algorithm, while the tumor classification models used the ResNet-101 algorithm.
Data set
The data set consisted of digital data scanned from MRI films representing patients from all over the country (most were from Southern China) who came to Sun Yat-sen University Zhongshan Ophthalmic Center (one of the most famous ophthalmic hospitals in China) for treatment. For all these patients, the diagnostic conclusions were supported by pathology and examined by the members of our team.
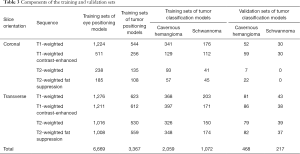
First, the MRI films brought by the patients from 45 different hospitals were scanned into a digital format and then screened, rotated and cropped. After this step, we obtained 6,507 images of cavernous hemangioma (from 33 different hospitals, Table 1) and 2,993 images of schwannoma (from 16 different hospitals, Table 2). Then, to form training sets and validation sets, we used the image processing software named LabelImg [Tzutalin. LabelImg. Git code (2015). https://github.com/tzutalin/labelImg] to interpret and manually label all the images. The purpose of interpretation is to generate coordinates that delineate the extent of the ranges of eyes and tumors according to anatomical knowledge. The labels include eye, cavernous hemangioma and schwannoma supported by pathological diagnosis. Next, all these processed data were randomly divided into two parts: a training set and a validation set. The training set included 6,669 images for the eye-positioning model, 3,367 images for the tumor-positioning model and 3,131 images (2,059 images for cavernous hemangioma and 1,072 images for schwannoma) for the classification model. The validation set included 468 images for cavernous hemangioma and 217 images for schwannoma (Table 3).

Full table

Full table
Full table
Experimental settings
The settings of this study were based on Caffe (18), the Berkeley Vision and Learning Center deep-learning framework (BVLC), and TensorFlow (19). All the models were trained in parallel on three NVIDIA Tesla P40 GPUs.
In terms of the classification problem, the key performance evaluation metrics were estimated as follows (20):
|
| [1] |
|
| [2] |
|
| [3] |
where N represented the quantity of samples; Pi represented the number of correctly classified samples within the ith class; k denoted the number of classes in this specific classification problem; TPi indicated the number of correctly classified samples within the ith class; FPi denoted the number of wrongly recognized samples within the ith class; FNi denoted the number of wrongly classified samples within the jth class,
For the object positioning problem, interpolated average precision (AP) was adopted for the performance evaluation (22). The interpolated AP is computed from the precision recall (PR) curve as shown in Eq. [4].
|
| [4] |
where
We adopted four-fold cross validation for the performance evaluation to assess all the classification and positioning problems.
Results
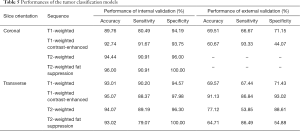
First, we conducted an internal four-fold cross-validation. The results showed that all the eye-positioning models achieved an AP of 100% and that the AP of the 28 tumor-positioning models exceeded 90% (Table 4). Similarly, the accuracy, sensitivity and specificity of almost all 32 tumor classification models were exceeded 90%, as shown in Table 5.

Full table
Full table
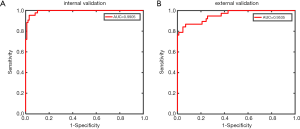
Next, we used the validation set for external validation. Considering that the tumor classification model results were mostly related to the differential diagnosis of cavernous hemangioma and schwannoma, the external verification of the tumor classification model predominantly represented the significance. The results showed that the transverse T1-weighted contrast-enhanced sequence model reached an accuracy of 91.13%, a sensitivity of 86.84%, a specificity of 93.02%, and an AUC of 0.9535. In contrast, the remaining models had significantly reduced performances compared with the internal verification results (see Table 5 and Figure 3).
Discussion
Good performance in a real-world setting
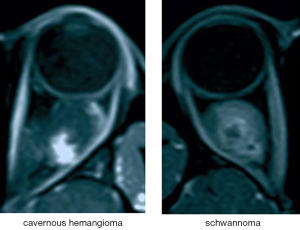
Based on clinical experience, T1-weighted contrast-enhanced sequences can highlight the blood vessels. Progressive filling from center to periphery on enhancement is typical of cavernous hemangioma, while the enhancement pattern of schwannoma is partial and uneven (5,6) (see Figure 4). Therefore, these sequences are considered the most significant reference among all types of slices in the differential diagnosis of the two studied tumor types (23,24). The tumor classification model trained by the transverse T1-weighted contrast-enhanced sequence images and tested on the external validation sets achieved high accuracy, sensitivity, and specificity in automated cavernous hemangioma and schwannoma differential diagnosis in a real-world setting that is completely consistent with the clinical environment.
Our results showed that the performance of the tumor classification model trained by transverse T1-weighted contrast-enhanced sequence images reached an accuracy of 91.13%, a sensitivity of 86.84%, a specificity of 93.02% and an AUC of 0.9535. These results suggested that this model’s performance quality meets the primary need for clinical application and that the goal of distinguishing cavernous hemangioma from schwannoma is achievable using this type of model.
A multicenter data-set
Thanks to the popularity of our ophthalmology center in China, patients from all over the country come here for treatment; thus, we were able to obtain these valuable images. In this study, we included data from more than 45 different hospitals in China to reach the current data amount. Moreover, due to the variety of equipment and operators among the different source hospitals, the data collection techniques were diverse, which enhances the wide generalizability of our diagnostic model.
Applying scanned versions rather than using DICOM
In previous AI studies, researchers have typically preferred raw data (11-13,15-17,25,26), such as DICOM format, generated directly from the imaging equipment, because the DICOM format both preserves all the original data and allows convenient collection. However, the scanned format was chosen for this study because the resultant AI framework needs to be useful for doctors in remote areas. The information technology level of hospitals in remote areas was limited, and they often lack comprehensive medical record management systems (27,28). Because most clinicians rely on film images instead of computerized interfaces, it made sense that models trained from a film version would be more suitable in this type of situation.
Three steps to reach the final goal
In previous studies, researchers commonly input entire MRI images for training (25,26). Here, we progressively designed three different types of models to achieve the goal of distinguishing cavernous hemangioma from schwannoma. First, because the eye area occupies only a small proportion of the entire MRI image, inputting the entire MRI image into the model directly would introduce considerable irrelevant information. To reduce the interference from such unnecessary information, we constructed an eye-positioning model that identifies the eye range within the full image; then, subsequent process can focus only on this range. Second, we established a tumor-positioning model to further narrow the scope for the final classifier and improve its precision. Third, we built a classification model to differentiate the located tumors to achieve the goal of automatically differentiating cavernous hemangioma from schwannoma.
Further subdividing the training sets instead of combining them
According to the traditional wisdom, having sufficient data volume is the foundation of training the current AI techniques (11-17). The most fundamental and effective way to improve the accuracy, sensitivity and specificity of the model is to augment the data in the training set. However, the MRI images for training had remarkable variations in different weighted sequences and slice orientations. If these images were blindly combined while ignoring these variations, the resultant incompatibilities would inevitably confuse the system, and its performance would deviate from the original intention. Therefore, we divided all the images into eight groups for training based on their different weighted sequences and slice orientations. The final result supported our conjecture: the performance of the transverse T1-weighted contrast-enhanced sequence was outstanding compared to that of other models. If all the training sets were combined, the accuracy of this model would be well below 91.13%.
Web-based automatic diagnostic system
Early in our research, our team built a cloud platform for congenital cataract diagnosis (29); we will implement the models in this study on that platform at the appropriate time. In China, an objective technological gap exists between urban and rural areas, and this imbalance is particularly evident with regard to medical resources (30-32). The establishment of this AI cloud platform for disease diagnosis is an economical and practical approach to alleviate the problem of the uneven distribution of medical resources.
Proper algorithms
Localization method
Faster-RCNN is a widely used algorithm used to address positioning problems because of its practicability and efficiency. Evolving from RCNN and Fast-RCNN (33), Faster-RCNN generates region proposals quickly by using an anchor mechanism rather than by applying a superpixel segmentation algorithm. After adopting two-stage training, transformations of the bounding box regressor and classifier were achieved. In the first stage, Faster-RCNN generated region proposals. Then, it judged the authenticity of the proposals, and the topmost coordinates of each object were regressed. In the second stage, the class of each object was evaluated and each object was eventually regressed to obtain its coordinates. We adopted a pretrained Zeiler and Fergus (ZF) network (34) to reduce the training time.
Convolutional neural network (CNN)
The CNN is the most popular AI model used in medicine. In this study, we adopted ResNet, which has a thin CNN architecture that includes numerous cross-layer connections and is suitable for rough classification tasks. To fit the residual function, the objective function was transformed, which resulted in a significant increase in efficacy, and we adopted a LogSoftMax loss function with class weights. The ResNet selected for this study has 101 layers, which is a sufficient depth to address the classification problems (20).
Limitations of our study
The most important deficiency in the study is that we simply chose a model that achieved good efficacy rather than also considering other models. Although the model trained on the group containing the transverse T1-weighted contrast-enhanced sequence images achieved particularly remarkable performance and is already sufficiently robust to help doctors in clinical work, the other seven groups may also contain useful information for feature extraction. Thus, the diagnostic efficiency of the model should be improvable to some extent if we were to make rational use of the other seven groups of data. Such an approach requires involving multimodal machine learning (35-37), because MRI images with different weighted sequences should be processed as separate modes. Upon alignment, the models could be integrated under the joint representation principle. Our team will continue to investigate this aspect of the problem in future studies.
Conclusions
The findings of our retrospective study show that the designed AI framework tested on external validation sets can achieve high accuracy, sensitivity, and specificity for differential diagnosis of automated cavernous hemangioma and schwannoma in real-world settings, which will contribute to the selection of appropriate treatments.
Although a partial accuracy rate of over 90% was achieved with the current data volume, AI algorithms can never have too much data. Thus, we plan to continue collecting additional cases to optimize the model by cooperating with hospitals in Shanghai to collect data in the eastern part of China, thereby supplementing our training set and enhancing model generalizability. Furthermore, at the appropriate time, we will design a web-based automatic diagnostic system to help solve the problem of obtaining advanced medical care in remote areas. In terms of algorithms, we will first investigate multimodal machine learning to take full advantage of these invaluable data. Overall, the results show that further investigation of AI approaches are clearly a worthwhile effort that should be tested in prospective clinical trials.
Acknowledgments
Funding: This study was funded by the National Key R&D Program of China (2018YFC0116500), the National Natural Science Foundation of China (81670887, 81870689 and 81800866), the Science and Technology Planning Projects of Guangdong Province (2018B010109008), and the Ph.D. Start-up Fund of the Natural Science Foundation of Guangdong Province of China (2017A030310549).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Haotian Lin and Limin Yu) for the series “Medical Artificial Intelligent Research” published in Annals of Translational Medicine. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.03.150). The series “Medical Artificial Intelligent Research” was commissioned by the editorial office without any funding or sponsorship. HL served as the unpaid Guest Editor of the series. The other authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Data collection, analysis, and publication of this study were approved by the Ethics Committee of the Zhongshan Ophthalmic Center (No. 2016KYPJ028) according to the principles of the Declaration of Helsinki.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shields JA, Bakewell B, Augsburger JJ, et al. Classification and incidence of space-occupying lesions of the orbit. A survey of 645 biopsies. Arch Ophthalmol 1984;102:1606-11. [Crossref] [PubMed]
- Tanaka A, Mihara F, Yoshiura T, et al. Differentiation of cavernous hemangioma from schwannoma of the orbit: a dynamic MRI study. AJR Am J Roentgenol 2004;183:1799-804. [Crossref] [PubMed]
- Shields JA, Shields CL, Scartozzi R. Survey of 1264 patients with orbital tumors and simulating lesions: The 2002 Montgomery Lecture, part 1. Ophthalmology 2004;111:997-1008. [Crossref] [PubMed]
- Scheuerle AF, Steiner HH, Kolling G, et al. Treatment and long-term outcome of patients with orbital cavernomas. Am J Ophthalmol 2004;138:237-44. [Crossref] [PubMed]
- Kapur R, Mafee MF, Lamba R, et al. Orbital schwannoma and neurofibroma: role of imaging. Neuroimaging Clin N Am 2005;15:159-74. [Crossref] [PubMed]
- Ansari SA, Mafee MF. Orbital cavernous hemangioma: role of imaging. Neuroimaging Clin N Am 2005;15:137-58. [Crossref] [PubMed]
- Calandriello L, Grimaldi G, Petrone G, et al. Cavernous venous malformation (cavernous hemangioma) of the orbit: Current concepts and a review of the literature. Surv Ophthalmol 2017;62:393-403. [Crossref] [PubMed]
- Young SM, Kim YD, Lee JH, et al. Radiological Analysis of Orbital Cavernous Hemangiomas: A Review and Comparison Between Computed Tomography and Magnetic Resonance Imaging. J Craniofac Surg 2018;29:712-6. [Crossref] [PubMed]
- Savignac A, Lecler A. Optic Nerve Meningioma Mimicking Cavernous Hemangioma. World Neurosurg 2018;110:301-2. [Crossref] [PubMed]
- Li Z, Keel S, Liu C, et al. An Automated Grading System for Detection of Vision-Threatening Referable Diabetic Retinopathy on the Basis of Color Fundus Photographs. Diabetes Care 2018;41:2509-16. [Crossref] [PubMed]
- Ting DS, Cheung CY, Lim G, et al. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images From Multiethnic Populations With Diabetes. JAMA 2017;318:2211-23. [Crossref] [PubMed]
- Gulshan V, Peng L, Coram M, et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016;316:2402-10. [Crossref] [PubMed]
- Gargeya R, Leng T. Automated Identification of Diabetic Retinopathy Using Deep Learning. Ophthalmology 2017;124:962-9. [Crossref] [PubMed]
- Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018;24:1559-67. [Crossref] [PubMed]
- Poplin R, Varadarajan AV, Blumer K, et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng 2018;2:158-64. [Crossref] [PubMed]
- Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. [Crossref] [PubMed]
- Li X, Zhang S, Zhang Q, et al. Diagnosis of thyroid cancer using deep convolutional neural network models applied to sonographic images: a retrospective, multicohort, diagnostic study. Lancet Oncol 2019;20:193-201. [Crossref] [PubMed]
- Jia Y, Shelhamer E, Donahue J, et al. Caffe: Convolutional Architecture for Fast Feature Embedding. arXiv:1408.5093 [cs.CV] 2014:675-8.
- Abadi M, Agarwal A, Barham P, et al. TensorFlow: large-scale machine learning on heterogeneous distributed systems. arXiv:1603.04467 [cs.DC] 2016:19.
- Zhang K, Liu X, Liu F, et al. An Interpretable and Expandable Deep Learning Diagnostic System for Multiple Ocular Diseases: Qualitative Study. J Med Internet Res 2018;20:e11144. [Crossref] [PubMed]
- Shi C, Pun C. Superpixel-based 3D deep neural networks for hyperspectral image classification. Pattern Recogn 2018;74:600-16. [Crossref]
- Everingham M, Zisserman A, Williams CKI, et al. The 2005 PASCAL Visual Object Classes Challenge. Berlin, Heidelberg: Springer Berlin Heidelberg, 2006:117-76.
- Jinhu Y, Jianping D, Xin L, et al. Dynamic enhancement features of cavernous sinus cavernous hemangiomas on conventional contrast-enhanced MR imaging. Am J Neuroradiol 2008;29:577-81. [Crossref] [PubMed]
- He K, Chen L, Zhu W, et al. Magnetic resonance standard for cavernous sinus hemangiomas: proposal for a diagnostic test. Eur Neurol 2014;72:116-24. [Crossref] [PubMed]
- Yasaka K, Akai H, Abe O, et al. Deep Learning with Convolutional Neural Network for Differentiation of Liver Masses at Dynamic Contrast-enhanced CT: A Preliminary Study. Radiology 2018;286:887-96. [Crossref] [PubMed]
- Lu Y, Yu Q, Gao Y, et al. Identification of Metastatic Lymph Nodes in MR Imaging with Faster Region-Based Convolutional Neural Networks. Cancer Res 2018;78:5135-43. [PubMed]
- Williams C, Asi Y, Raffenaud A, et al. The effect of information technology on hospital performance. Health Care Manag Sci 2016;19:338-46. [Crossref] [PubMed]
- Li H, Ni M, Wang P, et al. A Survey of the Current Situation of Clinical Biobanks in China. Biopreserv Biobank 2017;15:248-52. [Crossref] [PubMed]
- Long E, Lin H, Liu Z, et al. An artificial intelligence platform for the multihospital collaborative management of congenital cataracts. Nat Biomed Eng 2017;1:0024.
- Zhang T, Xu Y, Ren J, et al. Inequality in the distribution of health resources and health services in China: hospitals versus primary care institutions. Int J Equity Health 2017;16:42. [Crossref] [PubMed]
- Anand S, Fan VY, Zhang J, et al. China's human resources for health: quantity, quality, and distribution. Lancet 2008;372:1774-81. [Crossref] [PubMed]
- Fan L, Strasser-Weippl K, Li JJ, et al. Breast cancer in China. Lancet Oncol 2014;15:e279-89. [Crossref] [PubMed]
- Ren S, He K, Girshick R, et al. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans Pattern Anal Mach Intell 2017;39:1137-49. [Crossref] [PubMed]
- Zeiler MD, Fergus R. Visualizing and Understanding Convolutional Networks. Cham: Springer International Publishing, 2014:818-33.
- Baltrusaitis T, Ahuja C, Morency LP. Multimodal Machine Learning: A Survey and Taxonomy. IEEE Trans Pattern Anal Mach Intell 2019;41:423-43. [Crossref] [PubMed]
- Atrey PK, Hossain MA, El Saddik A, et al. Multimodal fusion for multimedia analysis: a survey. Multimedia Syst 2010;16:345-79. [Crossref]
- Ramachandram D, Taylor GW. Deep Multimodal Learning: A Survey on Recent Advances and Trends. IEEE Signal Proc Mag 2017;34:96-108. [Crossref]




