COVID-19 will stimulate a new coronavirus research breakthrough: a 20-year bibliometric analysis
Introduction
Coronavirus is an enveloped positive-sense single-stranded RNA virus. Its diameter is approximately 80 to 120 nm. It has the largest genetic material among all RNA viruses. It can infect humans, mice, cats, dogs, birds and other vertebrates (1-3). Coronaviruses have proliferated many times throughout the world, causing unimaginable harm to humanity. Words such as SARS and MERS have produced great fear in people’s hearts (4,5). There is no doubt that coronavirus has become a problem in the medical profession and even in society. However, with the COVID-19 outbreak in China, coronaviruses have once again become a focus (6).
COVID-19 is a new coronavirus strain that has never been found in humans before, and it is the seventh known coronavirus that can infect humans. It was discovered in a case of Wuhan viral pneumonia in 2019 and was named by the WHO on January 12, 2020 (7,8). As of this study, more than 100,000 people have been diagnosed with infection, and people of all ages can be infected. It has been confirmed that COVID-19 has the characteristics of human-to-human transmission and high concealment (7,9). Additionally, it has multiple transmission routes, including droplets, contact, and even aerosols, and the faecal-oral route may be included (10). Faced with this situation, to defeat the virus, there is still much work to be done by scientists in China and around the world.
In recent years, bibliometric analysis has become popular, which applies literature metrology characteristics to measure the contribution of an area of research, predicts detailed trends of research or hotspots in a certain field, and makes an important contribution to the prevention and treatment of diseases. However, there have been few bibliometric studies on coronaviruses, mainly focusing on MERS, and there is a lack of comprehensive analysis and research hotspot prediction for coronaviruses (11-13). In this article, we applied an integrated analysis of the content and external features of the research literature to summarize past research on coronaviruses and predict future research hotspots. We also provide an in-depth analysis of COVID-19 and summarize all the documented clinical trials to aid clinical treatment and scientific research.
Methods
Data sources and search strategies
Obviously, the Science Citation Index-Expanded and the Social Science Citation Index of Thomson Reuters’ Web of Science must be the most appropriate databases to perform bibliometric analysis. We searched Web of Science database comprehensively from 2000 to 2020, and only original articles and reviews were included. The search strategy was presented as follow: TI = (coronavirus) AND Language = English. To avoid bias cursed by frequent database renewal, all the literature retrieval and data download were completed in a single day February 9, 2020.
Data collection
Two reviewers (ZT and SZ) independently performed the primary search and their agreement rate reached 0.90, showing a significant accordance (14). WoSCC data including titles, countries, institutions journals authors and so on, were extracted and imported into the Online Analysis Platform of Literature Metrology (http://bibliometric.com/), CiteSpace V5.5.R1 SE, 64bit (Drexel University, Philadelphia, PA, USA) and VOSviewer (Leiden University, Leiden, the Netherlands) for bibliometric analysis. And the clinic trials data was obtained from ClinicalTrials.gov (https://clinicaltrials.gov/).
Bibliometric analysis
We tried to describe all publication characteristics, including countries, institutions, journals, authors, H index, and so on. We inquired the 2018 version of JCR (Journal Citation Reports) to get the impact factor (IF), which was regarded as an important indicator to measure the scientific value of research (15). In our study, we analyzed the annual publication numbers and growth tendencies of different country/region through Literature Metrology online analysis platform. CiteSpace is an optimal means for collaboration network analysis to connect all kind of publication characteristics. It can also obtain keywords with high citations to predict the research frontiers and emerging trends in this area. CiteSpace can apply “time slicing” function, for example, if you set the “years per slice” to one while the “top N per slice” is set to fifty, and the top fifty papers in a year would be exported into a single file. According to our objective, nodes of different size represented citation counts or publication counts (16,17). In addition, VOSviewer can sort keywords into different clusters based on co-occurrence analysis, and color them at the same time according to time course.
Results
Contribution of countries and institutions to global publications
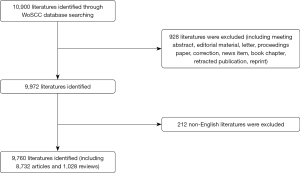
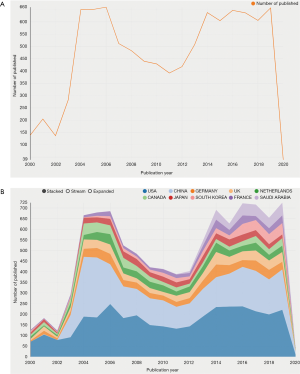
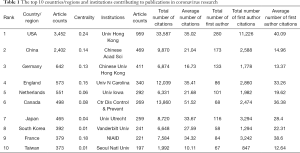
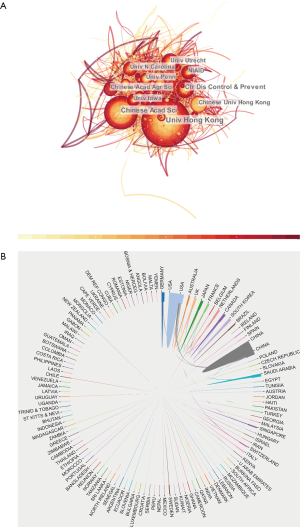
A total of 9,760 studies (8,732 articles and 1,028 reviews) met our inclusion criteria from 2000 to 2020 (Figure 1). Figure 2A displays a transformative trend in the annual literature numbers related to coronaviruses. All of the incorporated literature on coronaviruses was contributed by at least 114 different countries or regions (Figure 2B). The United States (n= 3,452) is the largest contributor to coronavirus research, followed by China (n=2,402), Germany (n=642), England (n=573), and the Netherlands (n=551). Centrality is a major indicator of the importance of nodes in a network, and a higher centrality means that a node is more important in a network, so the results showed that the United States has the most impact on other countries (centrality =0.24), followed by France (0.18) and England (0.15) (Table 1). In terms of research institutions, the top 10 include the University of Hong Kong (n=959), Chinese Academy of Sciences (n=469), Chinese University of Hong Kong (n=411), University of North Carolina (n=340), and University of Iowa (n=292) (Table 1). The coronavirus research network produces a low-density map (density =0.017) (Figure 3A), which means that the research teams are relatively scattered in various institutions, and increased mutual cooperation is needed. Most of the centrality indexes are below 0.15, indicating that the effect of most institutions stays at a low level and that the cooperation between institutions is insufficient. International cooperation analysis shows that the most frequent cooperation occurs in the United States and China (Figure 3B).
Full table
Journals publishing research on coronaviruses
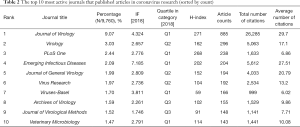
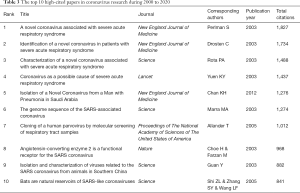
Recently, 1,323 journals have published research in the coronavirus field. The top 10 popular journals published 2,621 of all 9,760 studies on coronaviruses (26.85%) (Table 2). Among them, the top 3 journals are the Journal of Virology, Virology and PLoS One, which account for more than 14.54% of all indexed literature. The highest IF belongs to Emerging Infectious Diseases (7.185), followed by the Journal of Virology (4.324) and Viruses-Basel (3.811). According to the JCR 2018 standards, 5 journals are classified as Q1, 2 journals as Q2 and 3 journals as Q3. An analysis of highly cited papers showed that the New England Journal of Medicine and Science have an incredible scientific impact on all scholars, and 6 of the top 10 highly cited papers were published in these two journals (Table 3).
Full table
Full table
Contribution of authors to coronavirus research
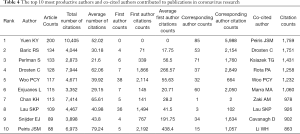
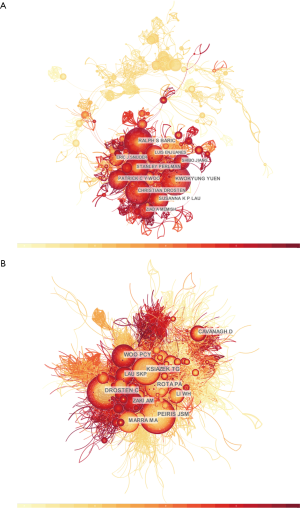
The ten authors that published the most papers, among all 29,515 authors, on this subject include Yuen KY, Baric RS, Perlman S, Drosten C, and Woo PCY (Table 4). Among them, Yuen KY, the chair of Infectious Diseases at the University of Hong Kong, ranks first, with 200 studies; Baric RS from the Department of Epidemiology, Program in Infectious Diseases, University of North Carolina at Chapel Hill in the USA is second, with 134 studies. These two scholars have made great achievements and become authorities in coronavirus research. We analysed the citation information of authors (Figure 4A) and co-cited authors (Figure 4B), visualizing it in a network produced by CiteSpace. Peiris JSM, with 1,759 co-citations, ranks first among the top ten co-cited authors, followed by Drosten C (n=1,751), Ksiazek TG (n=1,431), and Rota PA (n=1,258) (Table 4).
Full table
Analysis of coronavirus research hotspots
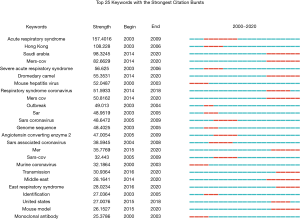
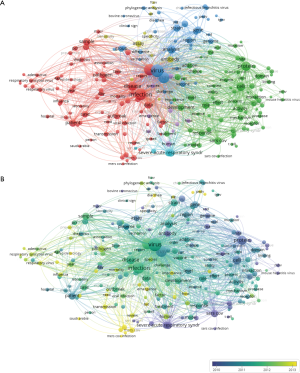
Keywords were extracted from 9,760 publications and analysed by VOSviewer. In Figure 5A, 216 keywords that appeared more than 200 times were included and classified into 5 clusters in the map: cluster 1 (clinical research, in red); cluster 2 (pathogenesis research, in green); cluster 3 (virological research, in blue); cluster 4 (treatment, in yellow) and cluster 5 (origin and transmission research, in purple). Circles with a large size represent the keywords that appeared at a high frequency. Within cluster 1, the following keywords frequently occurred: study (4,070 times), infection (4,057 times), disease (2,462 times), sample (1,672 times) and patient (1,641 times). In cluster 2, relevant keywords included protein (2,653 times), cell (2,381 times), role (1,575 times) and activity (1,393 times). In cluster 3, the primary keywords were virus (4,810 times), coronavirus (3,715 times), analysis (2,194 times), gene (1,624 times) and strain (1,562 times). Similarly, in cluster 4, the main keywords were antibody (1,207 times), assay (1,165 times), specificity (477 times), sensitivity (449 times) and evaluation (383 times). In cluster 5, they were human (920 times), species (719 times), identification (703 times), approach (659 times) and host (620 times). Detailed consequences of keywords are provided in Table S1. In Figure 5B, all keywords were coloured according to the average time of word appearance, from blue to yellow, representing early to recent appearances, respectively. We analysed the temporal trend of research hotspot shifts according to the top 25 keywords with the strongest citation bursts from 2000 to 2020 (Figure 6).
Full table
Discussion
Our statistical and quantitative analysis showed that the research output on coronavirus has fluctuated in the last 20 years. In Figure 2A,B, it can be seen that there was an explosion of research in this area during 2003–2006, with China and the United States contributing the most. There is no doubt that this increase is attributable to SARS in 2003. During that disaster, more than 5,000 people were infected with SARS coronavirus, including many medical staff, which caused massive panic worldwide. At that time, many scientists performed a multitude of research in this field, but after that, research on coronaviruses gradually decreased until 2012, when the outbreak of MERS caused research on coronaviruses to reach its peak again.
Regarding the contributions of countries and institutions, both the United States and China have played an important role in coronavirus research, and their total numbers of studies rank first and second, respectively. The United States seems to have superior conditions for basic medical research or clinical trials, which include adequate funding, advanced equipment and professional researchers. All the characteristics also show that the United States is leading the field. However, three institutions from China (the University of Hong Kong, Chinese Academy of Sciences and the Chinese University of Hong Kong) are ahead of scientific agencies in other regions. This phenomenon is partly because China was the main place where SARS occurred, and it also shows that the strength of scientific research from China has continuously increasing in recent years. The largest current problem is insufficient cooperation between various countries and institutions, which greatly reduces the efficiency of research. If there is improved communication and cooperation between institutions in various countries, I believe that research on viruses and diseases will achieve an enormous breakthrough.
Notably, the Journal of Virology published 885 studies in this area, far ahead of other journals. Other journals, including Virology, PLoS One and Emerging Infectious Diseases, were the primary journals containing coronavirus publications. In addition, the New England Journal of Medicine and Science focused on coronavirus research, and many highly cited papers were published in them. Thus, these findings imply that future developments in the field may be published in the aforementioned journals. Additionally, authors such as Yuen KY, Baric RS, Perlman S, and Drosten C not only published the largest numbers of papers in this field but also published their own highly cited representative papers in top magazines. Obviously, this publication record demonstrates that they have become an influential core group in the coronavirus field, having carried out substantial research to lay a solid foundation for future development.
We identified five keyword clusters to analyse research hotspots on coronaviruses. We found that the study of coronaviruses is relatively comprehensive, including clinical research, pathogenesis research, virological research, origin and transmission research and disease treatment method research. For research on coronavirus, we first need to understand its infectious disease characteristics, including its origin, susceptible population and transmission route, and then analyse its pathogenic mechanism and viral gene sequence to further find effective treatments and start clinical trials. In addition, the temporal trend of research hotspot shifts showed that the research in this field transferred mainly from early SARS to later MERS, which suggests that the increase in these studies was accompanied by the emergence of research hotspot events. The research increase had a very obvious time lag, which made us unprepared to deal with the emergencies. Therefore, we need to pay constant attention to various coronaviruses and their variants to prevent the emergence of large-scale infectious diseases.
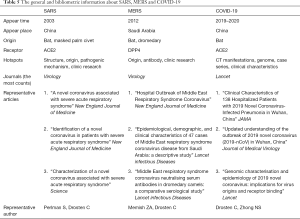
For COVID-19, although there are still few articles, we still summarized many of its bibliometric characteristics and compared them with those of SARS and MERS after our collection and analysis (Table 5). COVID-19 has similarity in gene sequence with SARS, and they have a common origin, bats, and a common intracellular receptor, ACE2 (18). Thus, the symptoms of COVID-19 are also similar to those of SARS, often manifesting as fever, cough, shortness of breath, or breathing difficulty, and in severe cases, pneumonia or even death may occur (19,20). However, COVID-19 is more concealed and more contagious than SARS. In the latest study from Guan et al., only 43.1% of patients had a fever when they were admitted to the hospital, and more patients had a fever during their hospital stay (21). For SARS or MERS, both of which are coronavirus induced, almost all patients have fever symptoms when diagnosed, and only 1–2% do not have a fever (22). This presentation means that if the screening of suspected cases relies only on measuring body temperature during epidemic prevention and control, then a large number of infected persons with no fever may be missed. After the Chinese Spring Festival, it is difficult to predict whether a second outbreak will occur as a large number of people return to work all over the country. Thus, this outbreak will be more difficult to address than SARS in 2003 (23).
Full table
Many scientists published a large number of articles after SARS occurred in 2003. Their main focus has been virus structure, origin, and pathogenic mechanism and clinical research. The journal Virology published the most literature, and there are some great studies published in top journals, such as the New England Journal of Medicine and Science. A similar situation occurred after the emergence of MERS. For COVID-19, although the number of articles is small, in a short period of time, many studies have been accepted by different top journals, such as JAMA and Lancet. Notably, we found that Drosten C has achieved great success in SARS, MERS and COVID-19 research (5,10). There is no doubt that he has become an authority in the field of coronavirus research. At this stage, CT manifestations, genomic sequence, and clinical characteristics are regarded as research hotspots in COVID-19 research, which is of great significance for the further prevention and control of disease. In addition, effective blockade of infectious pathways is also very important for disease prevention, and people now pay much attention to blocking droplets and air, but commonly touched objects cannot be ignored, such as elevators and shoes. Finally, we searched ClinicalTrials.gov and found 18 documented clinical trials (Table 6). Oxygen therapy, mechanical ventilation, empirical use of antibiotics and oseltamivir antivirals are the main methods currently used. Remdesivir and chloroquine, which have high potential, have been used in the clinic; although symptoms have improved, the therapeutic effects and side effects need further clinical trial verification. Immunoglobulin infusion, ECMO and other methods also have a certain effect on severe patients, and Chinese medicine should also be a good consideration for patients with different durations of infection. However, what is currently important is the development of new therapeutic drugs and vaccines, which may play a decisive role in defeating COVID-19. I believe that these clinical trials will provide reliable support for clinical research and have great guiding significance for the formulation of future therapeutic schedules.
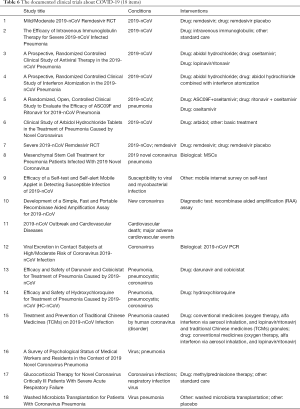
Full table
Nonetheless, some limitations may be inevitable. The database updates continuously, and we selected only the literature from 2000 to February 9, 2020, without literature published after that day. Therefore, there is a discrepancy between our bibliometric analysis and real publication conditions. The number of coronavirus studies may increase rapidly with the breakthrough of future research.
Conclusions
We assessed the publication information regarding different countries, institutions, authors, journals, etc. and analysed the research hotspots in the coronavirus field over the past 20 years based on these studies. COVID-19 must become the focus of coronavirus research in the near future. In addition, reviewing previous coronavirus studies and determining their similarities and differences with those on COVID-19 will help us to understand this new virus as soon as possible. Finally, clinical research on coronaviruses, especially randomized controlled trials, has great potential to guide the prevention and treatment of coronaviruses in the future. We believe our research can reflect novel directions for coronavirus research and help the Chinese people overcome this epidemic soon.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.04.26). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ziebuhr J. Molecular biology of severe acute respiratory syndrome coronavirus. Curr Opin Microbiol 2004;7:412-9. [Crossref] [PubMed]
- Sawicki SG, Sawicki DL, Siddell SG. A contemporary view of coronavirus transcription. J Virol 2007;81:20-9. [Crossref] [PubMed]
- Hagemeijer MC, Rottier PJ, de Haan CA. Biogenesis and dynamics of the coronavirus replicative structures. Viruses 2012;4:3245-69. [Crossref] [PubMed]
- Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019;17:181-92. [Crossref] [PubMed]
- Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003;348:1967-76. [Crossref] [PubMed]
- Wang C, Horby PW, Hayden FG, et al. A novel coronavirus outbreak of global health concern. Lancet 2020;395:470-3. [Crossref] [PubMed]
- Zhu N, Zhang D, Wang W, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 2020;382:727-33. [Crossref] [PubMed]
- WHO. Global research and innovation forum to mobilize international action in response to the novel coronavirus (2019-nCoV) emergency. 2020.
- Chan JFW, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020;395:514-23. [Crossref] [PubMed]
- Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med 2020;382:970-1. [Crossref] [PubMed]
- Bonilla-Aldana DK, Quintero-Rada K, Montoya-Posada JP, et al. SARS-CoV, MERS-CoV and now the 2019-novel CoV: Have we investigated enough about coronaviruses? - A bibliometric analysis. Travel Med Infect Dis 2020;33:101566. [Crossref] [PubMed]
- Wang Z, Chen Y, Cai G, et al. A Bibliometric Analysis of PubMed Literature on Middle East Respiratory Syndrome. Int J Environ Res Public Health 2016;13:583. [Crossref] [PubMed]
- Zyoud SH. Global research trends of Middle East respiratory syndrome coronavirus: a bibliometric analysis. BMC Infect Dis 2016;16:255. [Crossref] [PubMed]
- Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977;33:159-74. [Crossref] [PubMed]
- Eyre-Walker A, Stoletzki N. The assessment of science: the relative merits of post-publication review, the impact factor, and the number of citations. PLoS Biol 2013;11:e1001675. [Crossref] [PubMed]
- Chen C. CiteSpace II: Detecting and visualizing emerging trends and transient patterns in scientific literature. J Am Soc Inf Sci Technol 2014;57:359-77. [Crossref]
- Chen C, Ibekwe-Sanjuan F, Hou J, et al. The structure and dynamics of cocitation clusters: A multiple-perspective cocitation analysis. J Am Soc Inf Sci Technol 2010;61:1386-409. [Crossref]
- Zhou P, Yang XL, Wang XG, et al. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv 2020. [Crossref]
- Jin YH, Cai L, Cheng ZS, et al. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Mil Med Res 2020;7:4. [PubMed]
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. [Crossref] [PubMed]
- Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv 2020. doi: https://doi.org/ [Crossref]
- Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet 2015;386:995-1007. [Crossref] [PubMed]
- Chen J. Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes Infect 2020;22:69-71. [Crossref] [PubMed]