New era with the genetic assessment for biliary tree cancers beyond the anatomical assessment alone
We congratulate Dr. Hang and colleagues for their recently published study entitled “Cholangiocarcinoma: anatomical location-dependent clinical, prognostic, and genetic disparities” in Annals of Translational Medicine. In this unique study, the authors investigated the clinical impacts of the anatomical location of cholangiocarcinoma (CCA) using the SEER and TCGA databases. They elucidated prognostic outcomes in patients with CCA were most favorable in the distal type, and less so in the perihilar and intrahepatic types [distal (dCCA) > perihilar (hiCCA) > intrahepatic (iCCA)], whereas postsurgical prognosis was slightly higher in iCCA than in hiCCA. Furthermore, prognosis-predictive genes barely overlapped among the anatomical location of CCA. They insist that anatomical location could be strong indicator of the therapeutic strategy for CCA. Although this study comprehensively explored imbalance in anatomical location-dependent CCAs, several limitations need to be considered. The retrospective nature of the study means that clinical variables are not comprehensive. There are different historic stages among the three groups. To fully understand Hang et al.’s results, we have to appreciate the background of this research. The percentage of localized disease in dCCA, hiCCA, and iCCA was 18.8%, 22.0%, and 26.9%, respectively, while distant stage was 30.1%, 30.3%, and 43.5%, respectively. Stage IV in the TNM classification (AJCC 7th edition) constituted the majority (53.2%) in the SEER database. These differences in background could contribute to the disparities in prognostic outcomes among dCCA, hiCCA, and iCCA. We would encourage Hang and colleagues to extend their findings of different prognostic outcomes among the three groups by conducting propensity score matching analyses.
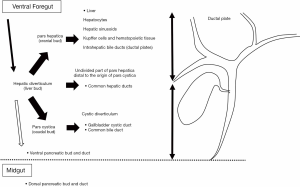
During human embryonic development, the intra- and extra-hepatic bile duct systems individually develops (1). The original anlage of the bile ducts and the liver is the hepatic diverticulum (liver bud), which enlarges rapidly and divides into two parts, namely the pars hepatica (cranial bud) and pars cystica (caudal bud). The pars hepatica gives rise to the liver, intrahepatic bile ducts and common hepatic duct. Its right and left branches become into the each of hepatic ducts. The distal side of the par’s hepatica forms the common hepatic duct. The proximal side of the main hilar ducts is derived from the intrahepatic ductal plate. On the other hand, the pars cystica gives rise to the cystic diverticulum, which in turn gives rise to the gallbladder cystic duct and common bile duct. Thus, the intrahepatic duct and main hilar duct originate from the cranial part of the hepatic diverticulum, whereas the extrahepatic duct develops through an extension of the caudate part of the hepatic diverticulum. Thus, the intra- and extra-hepatic biliary tree systems individually develop, and thereafter the two biliary systems join up. The junction of the foregut and midgut is at the opening of the common bile duct (Figure 1). Therefore, a common hepatic duct includes the boundary of the intrahepatic and extrahepatic biliary trees, suggesting that anatomical hilar CCAs include elements of both intrahepatic and extrahepatic biliary systems. Additionally, the intrahepatic small bile ducts are derived from hepatic progenitor cells via ductal plates. Recent cell-of-origin analysis by genetic assessment has elucidated that anatomical iCCAs may have different cell origins (features of hepatocytes or epithelial cells) derived from hepatoblasts (2). Wardell et al. (2) have reported that the most common classifications in 39 biliary tree cancer samples analyzed by whole-genome sequencing were liver (33.3%) and epithelial cell types (35.9%). iCCA samples were mainly originating from hepatocytes than those from epithelia (43.5% and 17.4%, respectively). Inversely, the other was true for dCCA samples (0% and 83.3%, respectively). Therefore, iCCA could be developed from the canals of Hering to the main bile duct, and is characterized as a heterogeneous CCA. Although the intrahepatic perihilar and distal bile ducts connect with each other and together form a biliary tree, the origins of these cells differ among iCCA, hiCCA, and dCCA. In summary, embryonic assessment in addition to anatomical assessment thus plays a crucial role in elucidating the genetic features of biliary tree cancers.
An alternative assessment of the real features of CCA is to elucidate the risk factors for biliary tree cancers. While most CCAs occur sporadically with no obvious cause, several risk factors such as chronic biliary and liver diseases, and liver flukes have been associated with the development of CCA (3). These various risk factors probably contribute to the wide geographical variation in the incidence of CCA (4). South East Asia has a high incidence of CCA. The major cause of CCA is due to infection by liver flukes such as Opisthorchis viverrini (Ov). From the genetic analyses of CCA with and without Ov infection, CCA with Ov was associated with upregulated genes involved in chronic inflammatory responses and xenobiotic metabolism, whereas CCA without Ov displayed enhanced expression of growth factor related genes such as HER2 (5). While CCA with Ov displays high incidence of the TP53 gene mutation, CCA without Ov is associated with the mutations of the IDH1, IDH2, and BAP1 genes (5). In contrast, CCA is relatively rare in Western countries. In Western countries, major risk factors of CCA are known as the primary sclerosing cholangitis, and choledochal cysts, and hepatolithiasis.
To determine the molecular mechanisms underlying CCA, it is thus important to recognize the background and etiology. The present study is based on the United States population, and the percentage by race was 79.1% White, 7.1% Black, and 13.8% Asian/others. Among the 45 CCA cases enrolled in TCGA, the background disease and race were 36 cases with iCCA, 7 with hiCCA, and 2 with dCCA and 38 White, 3 Black, and 4 Asian/others, respectively. Validation is required before Hang et al.’s results in can be applied to patients from other regions.
The features of CCAs thus depend on complex factors including embryonic and risk factors. On the other hand, Jusakul et al. (6) reported that anatomical sites of CCAs do not affect their survival trends, whereas molecular clusters elucidate remarkable differences in survival, in both their original and validation cohort. Jusakul et al. propose that assessment for molecular CCA subtypes in addition to anatomical location enhances to comprehend the cell biology and clinical behavior. In the present study, Hang et al. have performed a genetic assessment for CCAs similar those in previous reports (6,7). In their study, the prognostic behavior of each CCA type is highly distinct, proposing that independent bundle approach is required for dCCA, iCCA, and hiCCA. They also identified novel genes to estimate prognostic behavior accorfding to anatomical location of CCA. For example, the top genes for prognostic estimation of overall survival (OS) varied greatly between iCCA and hiCCA. In their analyses, overlapping genes in prognostic estimation among iCCA and hiCCA were extremely limited: PRLHR in OS for both iCCA and hiCCA, and SYNPR-AS1 in relapse-free survival (RFS) for hiCCA and OS for iCCA. Excluding pseudogenes and unknown genes, the top genes for OS were KCNMB2, NPFFR2, RIMBP2, SLC18A1 and VIT for iCCA, but CA9, DLX6, KRT20, MIR122HG, NRSN1, PLPR1, PLCXD3, RF00019, SLC13A1, and SLCO1B1 for hiCCA. For RFS, CYP3A43, AL590365.1, MIR200B, and AC109809.1 were most highly effective in iCCA, whereas AQP5, ASGR2, EVPL, HOXC10, MSLN, MUC16, NEFL, UGT1A10, and TMEM151A were highest in hiCCA.
Hang and colleagues’ comprehensive assessment for valuable genes in CCA provides clues to the molecular basis of biliary cancer development and progression. Genome-wide analysis of gene expression in CCA has recently become a useful tool for understanding molecular mechanisms and developing therapeutic strategies for patients with CCA. Several such analyses have been performed in iCCA (8,9) and biliary cancers (6,7), and have identified promising target genes for biliary cancers. An emerging molecular target in iCCA is mutant IDH1. IDH1 is a NADP(+)-dependent metabolic enzyme that catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate. IDH1 mutations induce a loss of function for oxidative decarboxylation of isocitrate, and a gain of function for the NADPH-dependent reduction of α-ketoglutarate to generate the oncometabolite D-2-hydroxyglutarate (10). Assessment of the frequency of mutant IDH1 in CCA subtypes suggests that it is higher in iCCA (11). The frequency of IDH1 mutation (mIDH1), recorded in 45 publications with a total of 5,393 patients, was 4,214 (78.1%) in iCCA, 1,123 (20.8%) in dCCA, and 56 (1.0%) in unknown anatomic locations (11). Borger et al. (12) reported that mutations in IDH1 and IDH2 were found only in iCCA (9 of 40, 23%) and in none of 22 dCCA and 25 gallbladder carcinomas. Thus, mIDH1 is a molecular feature of iCCA. Ivosidenib (mutant IDH1 inhibitor) has been approved to treat patients with refractory or relapsed acute myeloid leukemia by the US Food and Drug Administration, and has also exibited anti-cancer activity in patients with mIDH1-positive CCA in a phase I trial (13). In ivosidenib-treated patients with CCA, 6- and 12-month progression-free survival were 38% and 20%, respectively; 56% of patients displayed stable disease and 5% a partial response (13). A global phase III clinical trial to investigate the anti-cancer effect of ivosidenib is underway in previously treated advanced mIDH1 CCA (ClincialTrials.gov NCT02989857). Integrative analysis of a large CCA cohort has uncovered a novel molecular clustering in the present study. The study by Hang et al. included CCA patients only in the United States population and relatively advanced cases in clinical cohort analyses. Inadequate assessment can lead to misunderstanding of the molecular mechanism. Future experimental investigations will be important to clarify the underlying mechanisms of their identified genes. Despite its limitations, this study provides valuable information regarding disparities in clinical behavior and prognostic genes according to the anatomical location of CCA.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.03.206). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Roskams T, Desmet V. Embryology of extra- and intrahepatic bile ducts, the ductal plate. Anat Rec (Hoboken) 2008;291:628-35. [Crossref] [PubMed]
- Wardell CP, Fujita M, Yamada T, et al. Genomic characterization of biliary tract cancers identifies driver genes and predisposing mutations. J Hepatol 2018;68:959-69. [Crossref] [PubMed]
- Banales JM, Cardinale V, Carpino G, et al. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol 2016;13:261-80. [Crossref] [PubMed]
- Kirstein MM, Vogel A. Epidemiology and Risk Factors of Cholangiocarcinoma. Visc Med 2016;32:395-400. [Crossref] [PubMed]
- Ito T, Sakurai-Yageta M, Goto A, et al. Genomic and transcriptional alterations of cholangiocarcinoma. J Hepatobiliary Pancreat Sci 2014;21:380-7. [Crossref] [PubMed]
- Jusakul A, Cutcutache I, Yong CH, et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov 2017;7:1116-35. [Crossref] [PubMed]
- Miller G, Socci ND, Dhall D, et al. Genome wide analysis and clinical correlation of chromosomal and transcriptional mutations in cancers of the biliary tract. J Exp Clin Cancer Res 2009;28:62. [Crossref] [PubMed]
- Obama K, Ura K, Li M, et al. Genome-wide analysis of gene expression in human intrahepatic cholangiocarcinoma. Hepatology 2005;41:1339-48. [Crossref] [PubMed]
- Yang W, Li Y, Song X, et al. Genome-wide analysis of long noncoding RNA and mRNA co-expression profile in intrahepatic cholangiocarcinoma tissue by RNA sequencing. Oncotarget 2017;8:26591-9. [Crossref] [PubMed]
- Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009;462:739-44. [Crossref] [PubMed]
- Boscoe AN, Rolland C, Kelley RK. Frequency and prognostic significance of isocitrate dehydrogenase 1 mutations in cholangiocarcinoma: a systematic literature review. J Gastrointest Oncol 2019;10:751-65. [Crossref] [PubMed]
- Borger DR, Tanabe KK, Fan KC, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 2012;17:72-9. [Crossref] [PubMed]
- Lowery MA, Burris HA 3rd, Janku F, et al. Safety and activity of ivosidenib in patients with IDH1-mutant advanced cholangiocarcinoma: a phase 1 study. Lancet Gastroenterol Hepatol 2019;4:711-20. [Crossref] [PubMed]