
Next-generation sequencing diagnosis of severe pneumonia from fulminant psittacosis with multiple organ failure: a case report and literature review
Introduction
Psittacosis (ornithosis) is a naturally occurring infectious disease caused by Chlamydia psittaci (C. psittaci) in humans, birds, and some mammals. Clinical manifestations may vary from mild to fulminant. Symptoms often suddenly appear 5–21 days after exposure to the pathogen and can involve multiple systems. C. psittaci is a rare cause (<1%) of community-acquired pneumonia (CAP) (1) and few clinically diagnosed cases have been reported because of the lack of rapid and accurate diagnostic methods. Here, we report the use of next-generation sequencing (NGS) of the pathogen for a case of a successful diagnosis and treatment of severe pneumonia due to fulminant psittacosis with multiple organ failure at our hospital in December 2018.
Case presentation
A 50-year-old man who was an employee of the Market Supervision Bureau, was admitted to the emergency department of our hospital on December 5, 2018 due to a cough with phlegm for a week and fever with shortness of breath for 4 days. He was transferred to the department of respiratory and critical care medicine on December 6, 2018.
The patient started coughing, with white sticky phlegm and a sore throat, 7 days ago due to a cold. He had an episode of fever, with a maximum body temperature of 39 °C, and was coughing yellow phlegm, with chest tightness, shortness of breath, and dizziness. No chills, chest pain, hemoptysis, abdominal pain, and diarrhea were reported. The patient went to our hospital for emergency treatment. A routine blood test showed that the white blood cell count was 8.98×109/L, with a neutrophil ratio of 90.7%. The levels of high-sensitivity C-reactive protein (hs-CRP), procalcitonin (PCT), and interleukin 6 (IL-6) were 185.61 mg/L, 7.93 ng/mL, and 239.3 pg/mL, respectively. The biochemistry profile showed the following levels of enzymes: creatine kinase (CPK), 2,554 U/L; CPK-MB isoenzyme, 10.62 µg/L; lactate dehydrogenase (LDH), 1,673 U/L; glutamic oxaloacetic transaminase (GOP), 513.4 U/L; and glutamic-pyruvic transaminase (GPT), 183.1 U/L. Emergency chest computed tomography (CT) showed right lower lung infection and a heterogeneous, significantly enlarged right lobe of the thyroid. The patient recently had black loose stools but no bloody stools. He had an increased frequency of urination with urgency but no significant changes in the body weight.
The patient reported a history of thyroid enlargement for many years but did not seek a diagnosis and treatment by medical professionals. Instead, he took orally an unknown Chinese medicine for more than a year. He had a history of black stools for more than a year, which was considered as an appearance caused by the color property of the Chinese medicine, therefore, he did not seek a diagnosis and treatment. The patient is a government employee for market surveillance but denied recent exposure to poultry. His family history was unremarkable.
The vital signs of the physical examination on admission were as follows: body temperature, 36.6 °C; pulse, 107 beats/min; breathing, 32 times/min; blood pressure, 132/97 mmHg; saturated oxygen in arterial blood, 85%; and fractional inspired oxygen concentration (FiO2) with non-invasive ventilation, 50%. The patient was alert and collaborated during physical examination. His pupils were equal, round, and reactive to light. An enlarged thyroid (class II) was observed. The patient presented with a pharyngeal swelling and without any tonsillar swelling. Coarse breath sounds could be auscultated in the bilateral lungs. Scattered wet rales could be auscultated in the right lower lung. His heart rate was 107 beats/min, with a normal rhythm. No pathological murmurs were auscultated in the area of each valve. No abdominal tenderness and rebound tenderness were obvious. No edema was noted in the lower limbs.
The following diagnoses were made on admission: sepsis, severe pneumonia, multiple organ dysfunction (respiratory failure, myocardial damage, impaired liver function), gastrointestinal bleeding, enlarged right thyroid (unknown cause), hypokalemia, hyponatremia, and hypochloremia.
The outcomes of auxiliary examination after the admission were as follows: the blood gas analysis showed pH of 7.479, partial pressure of carbon dioxide of 29.3 mmHg, partial pressure of oxygen of 51 mmHg, base excess of −2 mmol/L, and FiO2 of 50%. A routine blood test showed that the platelet count was 94×109/L, and the neutrophil ratio was 92%. The biochemistry profile showed the following levels: blood urea nitrogen (BUN), 84 µmol/L; creatinine (Cr), 84 µmol/L; hs-CRP, 219.85 mg/L; PCT, 12.96 ng/mL; CPK 18,821 U/L; LDH, 1,622 U/L; GOP, 395.5 U/L; and GPT, 181.4 U/L. A routine urine test showed the urinary protein of 3.0 g/L and the red blood cell count of 200/µL. The coagulation function, hepatitis B surface antigen, syphilis antibody, HIV antibody, and hepatitis C antibody were within normal limits. A throat swab was performed and tested negative for the influenza virus antigen and bird flu nucleic acid. Smears stained for bacteria, fungi, and acid-resistant bacteria were negative in multiple tests. Bronchoscopy was performed, and the sputum smear was tested negative. Multiple blood cultures were negative. A urine culture test showed that the bacterial colony count was <100 colony-forming units per milliliter. Tests for the Mycoplasma pneumoniae antibody, Streptococcus pneumoniae antigen, blood and bronchoalveolar lavage fluid (BALF) galactomannan (GM) test, blood 1-3-β-D-dextran (G test), and cryptococcal antigen, as well as a blood tuberculosis immunoassay, were negative. The TORCH panel showed that the rubella virus IgG antibody was 34.9 IU/mL and the cytomegalovirus (CMV) IgG antibody was 189.3 AU/mL, while toxoplasma IgG, the IgM antibody, rubella virus IgM antibody, CMV IgM antibody, and herpes simplex virus (I + II) IgM were negative. Quantification of the blood CMV nucleic acid produced negative results, and the fecal occult blood test was negative.
Diagnosis and treatments
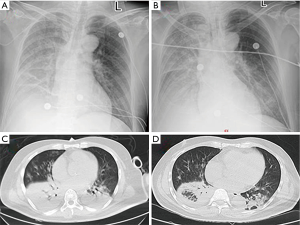
On the day of admission, a chest radiograph showed right lung infection and right pleural effusion (Figure 1A). 17 hours after the admission, the patient underwent endotracheal intubation with mechanical ventilation. He was administered imipenem/cilastatin, combined with linezolid and oseltamivir, for the treatment of infection and was also treated with methylprednisolone (80 mg daily). The patient’s condition was worsening rapidly. The chest radiograph was repeated 24 hours after the admission (Figure 1B) and showed decreased radiolucency in the right lung, patchy and large-patch shadows in the left middle and lower lung, which indicated aggravation of the lung lesion, and a right pleural effusion. Acute renal failure occurred 48 hours after the admission. The levels of BUN, Cr, PCT, lactic acid, IL-6, CPK, and CK-MB were 13.43 mmol/L, 210 µmol/L, 35.57 ng/mL, 2.13 mmol/L, 2,048 pg/mL, 13,371 U/L, and 160 U/L, respectively. On the 3rd day after admission, the patient underwent bronchoscopy. BALF and blood samples were obtained for NGS. On the 5th day after admission, the NGS results showed the presence of C. psittaci, and the reads of sequences detected in the BALF and blood samples were 137 and 1,134, respectively. Chest CT was repeated and showed aggregated lung infection and increased right lower lobe consolidation (Figure 1C); therefore, the antibiotics were changed to meropenem with oral doxycycline. On the 10th day after admission, the antibiotics were adjusted to doxycycline in combination with ceftazidime. On the 11th day after admission, the patient was weaned off mechanical ventilation. Tracheal intubation was removed, and the antibiotics were changed to oral doxycycline. On the 14th day after admission, chest CT was repeated and showed that the vague patchy shadows in the lungs improved, and a small amount of pleural effusion was noted on the right side (Figure 1D). A repeated biochemistry profile showed BUN of 4.3 mmol/L, Cr of 47 µmol/L, PCT of 0.15 ng/mL, lactic acid of 1.7 mmol/L, IL-6 of 9.1 pg/mL, CPK of 99 U/L, CK-MB of 22.4 U/L, GOP of 28 U/L, and GPT of 48 U/L. The patient was discharged on the 21st day after the admission. The diagnosis on discharge included severe pneumonia (C. psittaci), acute respiratory distress syndrome (ARDS), sepsis, multiple organ dysfunction (respiratory failure, myocardial damage, impaired liver function, and acute kidney injury), and euthyroid sick syndrome. We followed up the patient after 3 months, but he refused to come back for a review as his physical condition is good.
Discussion
Discussion among physicians from Shenzhen People’s Hospital, Shenzhen Institute of Respiratory Diseases
Psittacosis (ornithosis) is a naturally occurring infectious disease caused by C. psittaci in humans, birds, and some mammals. C. psittaci is a gram-negative obligate intracellular parasite, and birds are the main host. Bird infections are usually asymptomatic. Most human infections are transmitted from infected parrots (budgies, cockatoos, and parrots). Pigeons, canaries, birds, ducks, and swans can also be sources of human infection. Human infection is rarely associated with horses, cattle, koalas, and cats (2). Recently, it has been reported that peacocks and chickens can also carry the pathogen and cause disease transmission. There are occasional reports of person-to-person transmission, but this route of transmission is not considered important (3). Sporadic cases were reported in Asian countries; however, the incidence rate is lower in Asia than in Europe and Australia. The difference may be due to a higher rate of pathogen carriers among European parrots than among Asian parrots (4). The epidemiology and incidence of psittacosis are difficult to confirm because routine detection methods lack the specificity and sensitivity. The increasing use of molecular diagnostic methods in psittacosis is expected to provide more knowledge of the epidemiology of the disease (5,6).
The main route of human infection is through the respiratory tract by inhaling animal’s excretion aerosol. After the pathogen is inhaled, C. psittaci first enters reticuloendothelial cells of the liver and spleen to proliferate and then enters the lungs and other organs via the bloodstream. Therefore, human psittacosis is a systemic infection, with dominating respiratory infection. Clinical manifestations may vary from mild to fulminant. Sudden symptoms usually appear 5–21 days after exposure to the pathogen. The most common manifestations are fever, severe headache, myalgia, and dry cough. Other symptoms include a change in consciousness, photophobia, hepatosplenomegaly, and pharyngitis (2). When the respiratory system is involved, psittacosis can be manifested as an ordinary upper respiratory tract infection, pneumonia, and ARDS. When the heart is involved, psittacosis can be manifested as culture-negative endocarditis, myocarditis, and pericarditis. C. psittaci can be detected in the aortic valve, mitral valve tissue, and the blood. Arterial embolism has been reported. When the kidney is involved, psittacosis can be manifested as interstitial nephritis and acute renal failure. When the skin is involved, the disease can be manifested as Horder’s spots, rose rash, polytype erythema, nodular erythema, or urticaria. When the blood system is involved, in the acute phase, the white blood cell count is usually normal or slightly decreased. The infection can develop into leukopenia, hemolytic anemia, or disseminated intravascular coagulation in a few cases. It has been reported in the literature that C. psittaci causes cranial nerve palsy, transverse myelitis, Guillain-Barre syndrome, reactive arthritis, pancreatitis, and thyroiditis (2,3,7). Fulminant psittacosis combined with multiple organ failure is rare. These patients may have no obvious special symptoms at an early stage, but respiratory failure may occur as the disease progresses. Mechanical ventilation may be required for treatment. Fulminant psittacosis may be accompanied by varying degrees of septic shock and cognitive function impairment, kidney and liver failure, and abnormalities in the blood system. In some cases, extracorporeal circulation and hemodialysis may be required for treatment (8). Our patient’s condition was worsening rapidly, and respiratory failure was aggravated at 17 hours after the admission. He underwent mechanical ventilation and suffered from multiple organ (heart, liver, and kidney) failure. This is an extremely rare case of severe fulminant psittacosis. Because NGS provided diagnostic clues at an early stage of the disease, the patient was treated without delay. After 11 days of mechanical ventilation, the patient was weaned off mechanical ventilation and discharged within 20 days after the admission. His prognosis was good. Eighty percent of patients have abnormal findings on chest radiographs, which show different degrees of exudation and consolidation. The most common sign is single-lobe consolidation in which an air bronchogram sign can be noted. Consolidation is more severe in the lateral lung field and appears as a fan-shaped lesion. CT may show ground glass-like lesions and thickened bronchial vessels (7).
To date, the diagnosis of psittacosis has been made based on clinical manifestations, with at least one of the following laboratory examinations: (I) detection of C. psittaci in a respiratory specimen; (II) a complement-binding assay or a microimmunofluorescence assay showing antibody titers at least four-fold higher than the upper limit of normal in duplicate serum samples; (III) a microimmunofluorescence test showing that the titer of the IgM antibody is ≥1:16. Pathogen culture is generally used as a supplement to other tests and requires a high-level biosafety laboratory, and thus, it is not a routine test in most laboratories. Many factors can interfere with the results of serological tests, including an acute phase of the disease and early use of antibiotics, which may lead to false-negative results. A cross-reaction may lead to false-positive results. Therefore, molecular biology methods are becoming popular. Nucleic acid amplification assays are considered more sensitive and specific than are serological antibody tests (9). The abovementioned traditional detection methods require the clinician to provide directions for detection based on the patient’s epidemiological history and clinical manifestations. However, patients without a clear contact history are common. In this case, the patient’s occupation is a market supervisor. His medical history revealed that he often worked at a wholesale market. Although the patient denied a history of exposure to poultry, it is likely that his infection was related to the market environment. In addition, non-specific clinical manifestations and his episode in the flu season did not provide clues for clinicians to consider the rare disease of psittacosis. Therefore, simply relying on traditional diagnostic methods may lead to a misdiagnosis, missed diagnosis, and a delay in treatment. Fortunately, the patient was diagnosed by detecting the pathogen via NGS. The changed regimen saved the patient who was critically ill at the time of admission. He was discharged from the hospital after 20 days of treatment. NGS is also known as deep sequencing. The method does not target a specific pathogen, but high-throughput sequencing of nucleic acids is directly performed in clinical samples. The data reported herein can be compared with those in a database to determine the pathogenic microorganisms contained in the sample. This approach can quickly and objectively detect more and more rare pathogenic microorganisms in clinical samples, does not require specific amplification, and has an advantage for the diagnosis of rare pathogenic bacteria in difficult cases.
Doxycycline is a preferred agent for the treatment of psittacosis. The symptom of fever subsides in most patients after 48-hour treatment. In fulminant cases, if an oral drug or drug intestine absorption is not good, intravenous doxycycline can be administered for 14 days. Minocycline is also widely used in the clinic. Macrolides are preferred agents for pregnant women and children under 8 years old. The effect of quinolones in the treatment of psittacosis is not clear, and they are not recommended as first-line drugs (2). In this patient, a short-term use of quinolones before the diagnosis was not effective.
Several issues regarding the pathogenic mNGS and pathogenesis of severe pneumonia were further discussed as follows
Question 1. Which common and rare pathogens should be considered in the pathogenesis of severe pneumonia?
Expert opinion 1: Dr. Pedro J Marcos
In severe community acquired pneumonia an etiologic diagnosis use to be obtained at 50–60% of the cases with ordinary diagnostic methods (10,11). At the point when a pathogen is distinguished, Streptococcus pneumoniae is the pathogen all the more as often as possible included, pursued by H. flu (10,11). Recent series has observed that viruses are the third most common cause of sCAP in the ICU mostly due to the increase in the incidence of influenza pneumonia specially in winter periods (12). However, atypical pathogens are not uncommon since up to a 20% of this patient had a pneumonia related with an atypical pathogen, specially Legionella spp (13,14).
Expert opinion 2: Dr. Pierre Tattevin
A broad range of pathogens have been associated with severe pneumonia. These include bacteria (Streptococcus pneumoniae, Legionella pneumophila, Staphylococcus aureus—especially those producing Panton-Valentine leucocidin, Mycoplasma pneumoniae, Chlamydophila psittaci (15), Enterobacteriales, and many others), viruses (influenza, Middle East respiratory syndrome coronavirus, etc.), and fungi (e.g., histoplasmosis, Pneumocystis jiroveci in the immunocompromised (16). Although exposure, comorbidities, clinical presentation, and radiological features may guide investigations, the large number of potential etiologies has many drawbacks: (I) a large proportion of severe pneumonia remains of unknown etiology, even in 2020, which implies that anti-infectious treatment, and infection control measures cannot be targeted to the culprit pathogen; (II) the cost of etiological investigation may be high, if comprehensive diagnostic tests are undertaken, with limited return on investment.
Question 2. How can the diagnostic value of pathogenic mNGS in pulmonary infectious diseases be assessed?
Expert opinion 1: Dr. Pedro J Marcos
Up to now and with the best available clinical diagnostics a contributory pathogen can be detected in only a third of patients with respiratory infections. This is due to the low sensitivity and time requirements of culture, and the limited number of microbes detectable by serologic and PCR assays (12,17).
In this setting headways in genome sequencing like mNGS hold guarantee for over-coming these analytic difficulties by managing culture-independent evaluation of microbial genomes from extremely little clinical examples (microliters volumes).
It also has been described that this outperformed culture-depend method, especially for the detection of Mycobacterium tuberculosis (MTB), viruses, anaerobes and fungi. Furthermore, mNGS is less affected by prior antibiotic exposure (18) and mNGS can offer an improved detection of pulmonary infectious pathogens in lung biopsy tissues, with potential benefits in speed and sensitivity (19).
Expert opinion 2: Dr. Pierre Tattevin
New diagnostic tools (20), such as NGS (21), appear promising. These culture-independent methods, based on removal of human genes, followed by amplification and sequencing of microbial communities, typically through 16S rRNA genes for bacteria, or whole genome, have dramatically developed over recent years, with advent in high-throughput sequencing, machine-learning algorithms, and bio-informatic pipelines (22). NGS provides comprehensive profiling of microbial communities with relative abundance, and diversity, even in patients pre-treated with broad-spectrum antibiotics. Turn-around time may be dramatically reduced with portable, point-of-care devices. Once restricted to pioneer research laboratories, these diagnostic tools may replace current tools in the near future, for the routine diagnosis of complex infectious diseases, including for bedside diagnosis.
Severe pneumonia may be one of these. Studies have demonstrated that metagenomic identification of severe pneumonia pathogens is feasible, and accurate, in tracheal aspirates of patients under mechanical ventilation (20,21). Moreover, NGS may bring additional information, such as bacterial DNA burden, or decreased community diversity, both associated with identification of bacterial pathogens in bronchoalveolar lavage fluid (23), or tracheal aspirates (21).
Question 3. How to interpret the results of pathogenic mNGS?
Expert opinion 1: Dr. Pedro J Marcos
Maybe one of the most important strength is that mNGS offers the possibility of fast pathogen identification without a prior hypothesis of the target, so this is especially important in clinical practice to detect an unsuspected pathogen. Although at this moment the technology is really promising there are several hurdles that should be resolved before this new technology can be ready to be implemented in clinical laboratories. Some of them are the standardization of a quality control, workflow validation, method standardization and data interpretation. Anyway, this is transitory and can be overwhelmed by quickly developing advancements.
Another important issue is that the incredible sensitivity of mNGS make them able to detect not only the pathogens but also the airway microbiome, and transcriptional biomarkers of the host’s immune response. So, in the future if we want to integrate the mNGS into clinical practice when managing respiratory infections, it will be necessary to develop an mNGS-based method that integrates host response and unbiased microbe detection (24).
Expert opinion 2: Dr. Pierre Tattevin
NGS may not yet be ready for prime time, and still suffer from significant limitations: (I) accessibility to NGS platform remains low in most sites, even in wealthy countries, due to cost, turn-around time, and technical requirement; (II) simultaneous screening for bacterial, viral, and fungal pathogens with NGS is technically complex; (III) further studies are required to demonstrate the correlation between identification of pathogens through NGS, and their role in the disease under investigation. Nevertheless, the case reported herein is a clear demonstration that NGS may allow early identification of a pathogen that clinical reasoning, and standard microbiological tests would have missed.
Conclusions
For severe pneumonia, when the effect of empirical treatment is poor, it is of great significance to clarify the etiology of the disease. Second generation gene sequencing, as a new generation of detection technology, can be regarded as a good method when traditional detection methods have no results.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Written informed consent was obtained from the patient for publication of this Case Report and any accompanying images.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hogerwerf L, De Gier B, Baan B, et al. Chlamydia psittaci (psittacosis) as a cause of community-acquired pneumonia: a systematic review and meta-analysis. Epidemiol Infect 2017;145:3096-105. [Crossref] [PubMed]
- Stewardson AJ, Grayson ML. Psittacosis. Infect Dis Clin North Am 2010;24:7-25. [Crossref] [PubMed]
- Rybarczyk J, Versteele C, Lernout T, et al. Human psittacosis: a review with emphasis on surveillance in Belgium. Acta Clin Belg 2020;75:42-8. [Crossref] [PubMed]
- Cong W, Huang S, Zhang X, et al. Chlamydia psittaci exposure in pet birds. J Med Microbiol 2014;63:578-81. [Crossref] [PubMed]
- Cadario ME, Frutos MC, Arias MB, et al. Epidemiological and molecular characteristics of Chlamydia psittaci from 8 human cases of psittacosis and 4 related birds in Argentina. Rev Argent Microbiol 2017;49:323-7. [Crossref] [PubMed]
- Rane V, Khailin K, Williams J, et al. Underdiagnosis of Chlamydia trachomatis and Chlamydia psittaci revealed by introduction of respiratory multiplex PCR assay with Chlamydiaceae family primers. Diagn Microbiol Infect Dis 2018;90:163-6. [Crossref] [PubMed]
- Beeckman DS, Vanrompay DJ. Zoonotic Chlamydophila psittaci infections from a clinical perspective. Clin Microbiol Infect 2009;15:11-7. [Crossref] [PubMed]
- Wichert A, Lukasewitz P, Häuser M, et al. ARDS in fulminant ornithosis and treatment with extracorporeal lung assist. Int J Artif Organs 2000;23:371-4. [Crossref] [PubMed]
- Nieuwenhuizen AA, Dijkstra F, Notermans DW, et al. Laboratory methods for case finding in human psittacosis outbreaks: a systematic review. BMC Infect Dis 2018;18:442. [Crossref] [PubMed]
- Walden AP, Clarke GM, McKechnie S, et al. Patients with community acquired pneumonia admitted to European intensive care units: an epidemiological survey of the GenOSept cohort. Crit Care 2014;18:R58. [Crossref] [PubMed]
- Vallés J, Diaz E, Martín-Loeches I, et al. Evolution over a 15-year period of the clinical characteristics and outcomes of critically ill patients with severe community-acquired pneumonia. Med Intensiva 2016;40:238-45. [Crossref] [PubMed]
- Jain S, Self WH, Wunderink RG, et al. Community-acquired pneumonia requiring hospitalization among US adults. N Engl J Med 2015;373:415-27. [Crossref] [PubMed]
- Cillóniz C, Ewig S, Polverino E, et al. Microbial aetiology of community-acquired pneumonia and its relation to severity. Thorax 2011;66:340-6. [Crossref] [PubMed]
- Arnold FW, Summersgill JT, LaJoie AS, et al. A worldwide perspective of atypical pathogens in community-acquired pneumonia. Am J Respir Crit Care Med 2007;175:1086-93. [Crossref] [PubMed]
- Gacouin A, Revest M, Letheulle J, et al. Distinctive features between community-acquired pneumonia (CAP) due to Chlamydophila psittaci and CAP due to Legionella pneumophila admitted to the intensive care unit (ICU). Eur J Clin Microbiol Infect Dis 2012;31:2713-8. [Crossref] [PubMed]
- Wunderink RG, Waterer GW. Clinical practice. Community-acquired pneumonia. N Engl J Med 2014;370:543-51. [Crossref] [PubMed]
- Zaas AK, Garner BH, Tsalik EL, et al. The current epidemiology and clinical decisions surrounding acute respiratory infections. Trends Mol Med 2014;20:579-88. [Crossref] [PubMed]
- Miao Q, Ma Y, Wang Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis 2018;67:S231-40. [Crossref] [PubMed]
- Li H, Gao H, Meng H, et al. Detection of pulmonary infectious pathogens from lung biopsy tissues by metagenomic next-generation sequencing. Front Cell Infect Microbiol 2018;8:205. [Crossref] [PubMed]
- Yang L, Haidar G, Zia H, et al. Metagenomic identification of severe pneumonia pathogens in mechanically-ventilated patients: a feasibility and clinical validity study. Respir Res 2019;20:265. [Crossref] [PubMed]
- Kitsios GD, Fitch A, Manatakis DV, et al. Respiratory microbiome profiling for etiologic diagnosis of pneumonia in mechanically ventilated patients. Front Microbiol 2018;9:1413. [Crossref] [PubMed]
- Leo S, Gaïa N, Ruppé E, et al. Detection of bacterial pathogens from broncho-alveolar lavage by next-generation sequencing. Int J Mol Sci 2017;18:2011. [Crossref] [PubMed]
- Dickson RP, Erb-Downward JR, Prescott HC, et al. Analysis of culture-dependent versus culture-independent techniques for identification of bacteria in clinically obtained bronchoalveolar lavage fluid. J Clin Microbiol 2014;52:3605-13. [Crossref] [PubMed]
- Langelier C, Kalantar KL, Moazed F, et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc Natl Acad Sci U S A 2018;115:E12353-62. [Crossref] [PubMed]


