
Non-disruptive mutation in TP53 DNA-binding domain is a beneficial factor of esophageal squamous cell carcinoma
Introduction
Esophageal cancer is one of the deadliest diseases worldwide, and 90% of esophageal cancer cases belong to esophageal squamous cell carcinoma (ESCC) in China (1,2). The tumor suppressor gene TP53 is the most frequently mutated gene in ESCC. This gene comprises 11 exons and 10 introns. The p53 protein encoded by TP53, is a 393 amino acid residue protein with seven functional domains, including an acidic N-terminus transcription activation domain (TAD) from residue 1 to 42 and 55 to 75, an activation domain 2 (AD2) from residue 43 to 63, a DNA-binding domain (DBD) from residue 102 to 292, a nuclear localization signaling (NLS) domain from residue 316 to 325, a C-terminal oligomerization domain (OD) from residue 307 to 355, and a tetramerization domain (TET) from residue 356 to 393 (3,4). The coding sequence of TP53 gene comprises five regions, namely, 13–19, 117–142, 171–192, 236–258, and 270–286, that show a high degree of conservation among vertebrates, primarily in exons 2, 4, 5, 7, and 8, respectively. Aside from the coding region 13–19, the other four conserved areas are located in the DBD (4-6). The p53 DBD provides a scaffold for a flexible DNA-binding surface, which is formed by two large loops (loop L2, residues 163–195; L3, residues 236–251) that bind to a zinc atom (7). The transcriptional activity mediated by the DBD is the primary mechanism underlying the tumor suppressor activity of p53 (8).
p53 plays a crucial role in many cellular processes, including autophagy (9), metabolism (10), differentiation (11), and DNA repair. It is one of the most commonly mutated genes in human cancers, and over 50% human tumors carry TP53 mutations (12,13). Mutant p53 has been reported to overturn crucial cellular pathways and promote cancer cell proliferation and survival, invasion, migration, metastasis, and chemoresistance (12-15). However, mutant p53 protein not only loses its tumor suppressive functions but also gains new oncogenic properties (16). The function and prognostic values of mutant p53 are yet incompletely understood (4,17).
Several criteria have been used to classify TP53 mutations, including mutation status, mutation number, allele frequency, mutation region, degree of disturbance in p53 protein structure or function, and p53 protein expression. Classification into “disruptive” and “non-disruptive” forms based on functional effects on p53 protein has been proposed (18). Disruptive mutations are defined as (I) any mutations that introduce a stop codon (nonsense, frameshift, and intronic) or (II) an in-frame deletion within the L2 or L3 loop or missense mutations in the L2 or L3 loop replacing one residue by another with different polarity or charge. Non-disruptive variations include (I) missense mutations and in-frame deletions outside the L2–L3 loop or (II) missense mutations within the L2–L3 loop without any change in polarity or charge (8,18). Disruptive mutations are likely to cause loss of activity of p53 protein, while non-disruptive variants may retain the functional properties of wild-type p53. Skinner and colleagues proved that disruptive TP53 mutations lead to locoregional recurrence in head and neck cancers (19). Non-disruptive mutation serves as an independent prognostic factor of shorter survival in advanced non-small lung cancer (8). Considerable efforts have been directed to clarify the impact of TP53 mutations on the prognosis of patients with ESCC, but the results remain controversial. The number of patients enrolled, differences in follow-up methods and time, and various classifiers of TP53 mutations have led to contradictory outcomes, particularly the scattered mutation spectrum of TP53 (20). ESCC is one of the lethal cancers, highlighting the need for the discovery of novel biomarkers to assist disease management (21).
Here, we examined the whole exons of TP53 gene in 161 patients with resectable ESCC by next-generation sequencing (NGS), and analyzed the expression level of p53 protein by immunohistochemistry (IHC). We stratified patients by multiple TP53 mutation classifiers and analyzed the correlation of TP53 mutations with clinical parameters. We identified the most relevant classification of TP53 mutations with respect to patient outcome.
Methods
Patients and samples
Formalin-fixed paraffin-embedded (FFPE) specimens with matched blood samples as reasonable controls were available from 161 patients with ESCC. These patients underwent surgery from May 2008 to June 2014, and their tissue samples were collected and stored in the Tissue Bank of Zhejiang Cancer Hospital. All subjects had provided written informed consent, and this study was conducted following the Declaration of Helsinki Principles and approved by the Institutional Review Committee of Zhejiang Cancer Hospital. Patient data were available for age, gender, body weight, height, smoking and alcohol consumption status, and tumor size, localization, differentiation, TNM stage, surgery, and treatment. The 8th edition of AJCC/UICC staging system was used for TNM staging. Information on tumor differentiation and histopathologic classification was collected from pathology reports and independently examined by two senior pathologists.
NGS and data analysis
FFPE samples containing at least 20% tumor cells [as determined from the examination of hematoxylin and eosin (H&E)-stained sections] were deparaffinized and genomic DNA (gDNA) was extracted using QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) in accordance with manufacturer’s instructions, followed by quantification using PicoGreen fluorescence assay (Invitrogen). The gDNA from white blood cell (WBC) samples was extracted using QIAamp DNA Blood Mini Kit (Qiagen) as described by the manufacturer.
All sequencing processes were accomplished in 3DMed Medical Laboratory Co., Ltd (Shanghai) (22). The details of NGS method are described in manuscript communicated for publication (Paper #NCOMMS-18-38299C). Illumina NextSeq 500 was used to sequence samples with the IDT xGen hybridization buffer. To evaluate the quality of the sequencing data, we used FastQC software (
Classification of TP53 mutations
Mutations were classified as “disruptive” and “non-disruptive”, as per a reported article (18). Supplementary Table S1 shows the other six summarized criteria, including TP53 mutation status, mutation numbers, mutation frequency, degree of disturbance of p53 protein structure or function, functional domain, and domain and function.
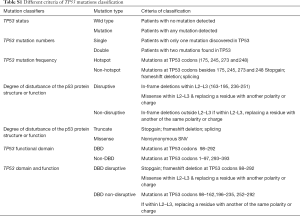
Full table
Assessment of IHC
Mouse anti-p53 protein monoclonal antibody (ZM-0408, ZSGB-BIO, Beijing, China) was used to detect the expression of p53 in FFPE specimens. Complete IHC protocols are described in our previous study (25). p53-stained slides were digitally imaged with a Digital slice scanner (KF-PRO-005-EX) and graded by two independent pathologists. Intensity was scored as 0 (no staining), 1 (weak staining), 2 (moderate staining), and 3 (strong staining) (26,27). p53 expression level in each sample was assessed as per IHC score, which was calculated using the following formula: staining intensity × percentage of positive cells (28-30). The resulting score ranged from 0 to 300. Receiver operating characteristic (ROC) curve analysis was performed to obtain the best cutoff values by the Youden index (sensitivity + specificity − 1) (31) to divide patients into two cohorts as follows: low expression and high expression.
Statistical analyses
Descriptive statistics were used to summarize the characteristics of patients; the results were expressed as frequencies and percentages for categorical variables. All factors were considered as categorical variables. Spearman’s rank correlation analysis was used to assess the correlation between TP53 mutation status and clinicopathologic features. Differences in the distribution of TP53 mutation types under various clinicopathologic variables were evaluated using the chi-square test.
Progression-free survival (PFS) was calculated for the patients in our ESCC cohort from time of surgery to cancer recurrence or last follow-up. Overall survival (OS) was defined as the time from surgery to death or last follow-up. The data for patients who were alive without recurrence at the time of analysis were censored at the last follow-up. Median PFS and OS and 95% confidence interval (CI) were evaluated using the Kaplan-Meier method, and survival curves were compared by the log-rank test. The Cox proportional hazard model was used to explore possible survival differences and identify factors affecting survival. Cox regression univariate and multivariate analyses were used to generate survival hazard ratio (HR) and 95% CI. Levels of statistical significance were bilaterally set at P<0.05. All calculations were performed with Statistical Package for Social Science (SPSS) for Windows (version 19.0; IBM Corp., Armonk, NY), and figures were created using GraphPad Prism (version 7.0; GraphPad Software, San Diego, CA).
Results
Patient characteristics and TP53 status
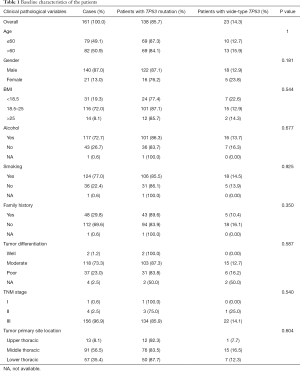
In total, 161 patients with ESCC were grouped according to TP53 mutation status as detected by NGS, and their clinicopathologic features are shown in Table 1. The median age of the cohort was 61 years and 50.9% patients were older than 60 years. In total, 87.0% were males. The majority of patients had smoking (77.0%) and drinking (72.7%) habits, and 8.1% patients were considered obese with a body mass index (BMI) >25. Based on pathological characteristics, 96.9% tumors were TNM stage III, 73.3% were moderately differentiated tumor, and 56.5% were located in middle thoracic. For treatment, 3.72% patients received neoadjuvant treatment, 49.07% received adjuvant treatment, and 47.2% [76] patients received neither neoadjuvant nor adjuvant treatment. TP53 mutations were detected in tumors from 138 patients (85.7%), and the mutation status was not significantly associated with gender, histology, BMI, smoking, alcohol consumption, family history, tumor stage, differentiation, or location in either TP53 wild-type (TP53-wt) or TP53 mutant (TP53-mut) group (Table 1).
Full table
Mutational landscape of TP53
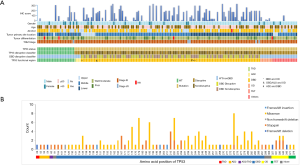
All coding exons of TP53 gene were examined by NGS, and 163 mutations were discovered in 138 patients. In general, 85.7% (138/161) patients had TP53 mutations. The different types of TP53 mutations detected in our study and their distribution are shown in Figure 1. Most patients (113/138, 81.9%) carried only one TP53 mutation, while 15.5% had double mutations. TP53 mutations were detected in exons 3 to 11, and were dominant among exons 5 to 8 (109/161, 67.7%) (Figure 1B). These mutations were mainly detected in DBD (111/161, 68.9%) (Figure 1A). Missense mutation was the most frequently detected mutation (97/163, 59.5%), followed by stop-gain (34/163, 20.9%), splicing (18/163, 11.0%), and frameshift deletion/insertion (8/163, 4.9%) (Figure 1B). The most frequently occurring variation was R273X (H/L/C) that accounted for 4.9% (appeared in 8 cases) cases, followed by Y220X (C/*) discovered in 7 patients, H193 (Y/L/R) and H179X (Y/L/R) in 6 patients, and R175H in 5 cases (Figure 1B). In addition, 55.8% (77/138) patients with TP53 mutations showed disruptive mutations, of which 64.9% (50/77) were observed in DBD (Figure 1A).
TP53 mutation classification and survival
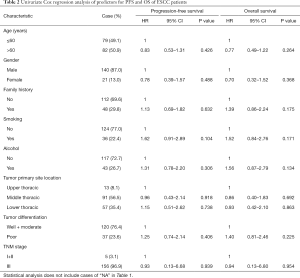
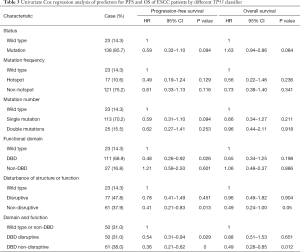
The follow-up period ranged from 0.1 to 120 months, with a median of 39.47 months for patients whose data were censored. During follow-up, 88 cases of recurrence and 87 deaths due to tumor progression were reported. Univariate Cox analysis showed that clinical pathological variables were not predictors of PFS and OS (Table 2). TP53-mut patients had a median OS of 25.57 months versus 38.35 months for TP53-wt patients, but the difference was not statistically significant (HR: 0.708; 95% CI, 0.37–1.34; P=0.29, Table 3). Different types of mutations in TP53 gene have different effects on the functionality of the protein. Hence, we stratified patients into multiple TP53 mutation classifiers based on different mutant features (Table S1). Some TP53 mutation classifiers, including hotspot mutations, mutation numbers, and allele frequency (data not shown), failed to predict the prognosis of patients (Table 3).
Full table
Full table
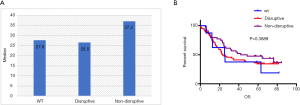
Mutations in DBD showed benefit in PFS (HR: 0.48, 95% CI: 0.26–0.92, P=0.026, Table 3, Figure 2A) but no significance with OS (HR: 0.65, 95% CI: 0.34–1.25, P=0.198, Table 3, Figure 2B). According to the degree of disturbance to the structure and function of p53 protein, we divided the mutations into two categories, namely the “disruptive” and “non-disruptive” type, and found that patients with non-disruptive mutation had better PFS (HR: 0.41, 95% CI: 0.21–0.83, P=0.013, Table 3, Figure 2C) and extended OS (HR: 0.49, 95% CI: 0.24–1.00, P=0.050, Table 3, Figure 2D). Together the results of DBD and disruptive analyses led to the creation of a new classifier, “DBD disruptive” and “DBD non-disruptive”. Univariate Cox regression analysis showed that the patients with non-disruptive p53 mutation in DBD had better PFS (P<0.001, Table 3, Figure 2E) and OS (P=0.005, Table 3, Figure 2F) than those with TP53-WT or TP53-mut not located in DBD.
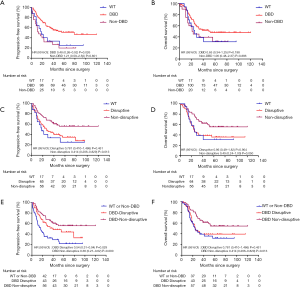
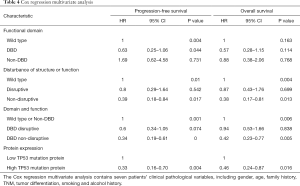
In the multivariate Cox proportional hazard model (Table 4), the presence of a DBD non-disruptive TP53 mutation was significantly associated with increased PFS (HR: 0.34; 95% CI: 0.19–0.61; P=0.000) and OS (HR: 0.42; 95% CI, 0.23–0.77; P=0.005). The presence of non-disruptive TP53 mutation in DBD was an independent prognostic factor for resectable ESCC.
Full table
IHC
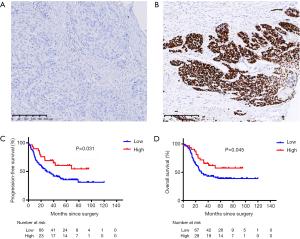
The IHC result was shown in Figure 3. The best cutoff value of 170 was used to distinguish patients into low and high p53 expression groups. Of these, 77.1% (118/153) patients were categorized into the low expression group and 35 into the high expression group. The median IHC score was 161.8, 106.1, and 89.2 in exon 7, 8, and 5, respectively. Exons 5–8 were the top 4 locations for mutations and mutated protein expression (Figure 1A). The results of chi-square test showed that the expression of p53 was associated with missense mutations (P<0.001), mutations in DBD (P=0.001), hotspot mutations (P=0.020), disruptive mutation (P=0.010), and non-disruptive mutation in DBD (P=0.001) (Table S2). The expression level of TP53 protein was independent of the mutational status (P=0.117) and mutation numbers (P=0.270). Furthermore, Cox regression univariate analysis showed that the patients from the high p53 expression group showed better outcomes (PFS: HR: 0.33, P=0.004; OS: HR: 0.46, P=0.016) (Table 3).

Full table
Conclusions
We analyzed TP53 mutations in 161 patients with resectable ESCC and described a new standard method to classify TP53 mutations. TP53 non-disruptive mutation located in DBD characterizes a distinct prognostic group of patients with ESCC with significantly extended survival. We found that patients with high p53 protein expression (IHC score >170) showed better outcomes. TP53 non-disruptive mutation in DBD and IHC results highlight the clinical usefulness of this prognostic marker in resectable ESCC.
We detected TP53 mutations in 85.71% patients with ESCC, consistent with the frequency described in The Cancer Genome Atlas (TCGA) database. However, different studies have shown variations in TP53 mutation frequency in ESCC, as determined by sequence coverage and other methods. Examination of exons 5 to 8 with traditional methods such as Sanger sequencing showed that almost 40% patients carried TP53 mutations (32-34). TP53 mutation frequency may reach up to 93% with NGS in ESCC (35). This phenomenon shows that the genomic region is essential for TP53 genotyping. The most frequently detected TP53 mutation type in ESCC was C>T transition (up to 85%) that was located in exons 5 to 8 (35). We found similar results. Nonsynonymous SNV was the most dominant mutation. In general, the TP53 mutational landscape observed in the present study is consistent with that previously reported.
Hotspot mutations are important for driver genes such as EGFR primarily located in exons 18–21. In such situations, target NGS panel, droplet digital polymerase chain reaction (PCR), or quantitative PCR instead of whole exome sequencing, may reduce the cost and turnaround time. However, TP53 mutations are dispersed in human cancers, and aside from the “hotspot mutations”, several other mutations are known to affect p53 protein functions. Hotspot mutations of TP53 are inconsistent in different studies. Maeng (36) found TP53 hotspot mutations in R306, R175H, and R273C, but others have defined hotspot mutations in R175, G245, R248, R249, R273, and R282 in ESCC (37,38). In our study, we found some variants, including R273X, Y220X, H179X, H193, and R175, that showed frequent mutations. However, these “hotspots” were not so frequent, as the most common mutation R273X appeared only in eight cases. Hence, it is much more suitable to detect TP53 gene by NGS instead of identifying hotspot mutations.
TP53 mutation, one of the most frequently observed mutations in human cancers, has been studied in various carcinomas (39). Studies with TP53 have mainly focused on mutation status and analyzed the effect of prognosis or clinical features, including smoking, drinking, and family history of cancer (40). However, recent reports have shown the shortcomings associated with these classifications. Efforts have been directed to define TP53 mutations to understand the exact nature of TP53. As per the effects on p53 protein function, Poeta and his colleagues (18) first proposed a standard method in head and neck squamous cell carcinoma (HNSCC) by dividing mutations into “disruptive” and “non-disruptive” forms. Matteo Canale (41) and colleagues tried to use a different exon mutation to classify TP53 mutations in non-small cell lung cancer (NSCLC). A meta-analysis showed that the OS of ESCC patients with different TP53 mutation number, frequency of allele was no differential in survival outcomes (21). Several studies have proved that the expression of p53 is more critical than TP53 mutations (21,42). Different mutation could result in different proteins, activate or suppress signaling pathways, and produce a range of significant biological effects (43,44). Hence, we considered the impact of risk factors for ESCC on prognosis, including BMI, gender, smoking, and alcohol consumption. However, we failed to observe any direct evidence that these risk factors would reduce PFS or OS.
Several strategies have been used to group TP53 mutations. After many attempts, we classified TP53 mutations into “disruptive” or “non-disruptive” types. This classification has been used with HNSCC (18), NSCLC (8), breast cancer (16), and ovarian cancer (45). However, no research report has described this classification in ESCC. In comparison with patients from disruptive mutation group, those from TP53 non-disruptive mutation group had better treatment response for head and neck cancer (19). However, in NSCLC, TP53 disruptive cluster showed prolonged OS (8). In our study, we clearly found that non-disruptive TP53 mutation was associated with good prognosis. In ovarian cancer, disruptive TP53 mutations showed survival benefits (45). The association between TP53 non-disruptive mutation and prognosis was significantly different in various cancers and may be related to the following factors: pathological types of tumors (adenocarcinoma versus squamous cell carcinoma) (42), treatment regime (new targeted therapy versus traditional radiotherapy/chemotherapy), and other molecular features. To test and verify our results, we used the whole exome sequencing data by Gao et al. (35) available at the European Genome-phenome Archive (EGA) under the accession number EGAS00001000932. It included results of 113 Chinese patients with ESCC. Even with a P value >0.05, a trend of non-disruptive mutation showing longer OS than the other two types was observed (Figure S1).
The result of IHC proves our view. p53 expression level and related mutations were associated with the prognosis of patients. IHC of p53 was related to some mutations, which affected protein expression.
In spite of the specificity and sensitivity of IHC and the overexpression of WT p53 (46,47), five samples considered as WT by NGS showed false-positive results. As p53 is a regular routine index in pathological IHC reports, the conversion of staining results into IHC scores is convenient. Hence, the use of this value to estimate prognosis in clinic may be valuable for patients that cannot afford sequencing and may help clinicians to access patient prognosis.
Some limitations of this study include the limited case numbers with TP53 WT and stage I and II cases and incomplete data (such as smoking and alcohol history did not distinguish between former consumers and non-consumers).
In conclusion, we demonstrate that the non-disruptive mutation in TP53 DBD and p53 expression level both have significant clinical importance in patients with resectable ESCC. These parameters may help clinicians to assess the prognosis of patients.
Acknowledgments
The authors thank Qianqian Yao, Wenting He and Meng Wang for their technical support and insightful discussions.
Funding: This study was supported by the National Natural Science Foundation of China (81472203 to S.D.); the Major Science and Technology Plan of Zhejiang Medicine and Health funded by the National Health Commission (WKJ-ZJ-1902 to S.D.)
Footnote
Conflict of Interests: The authors declare no conflict of interest.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All subjects had provided written informed consent, and this study was conducted following the Declaration of Helsinki Principles and approved by the Institutional Review Committee of Zhejiang Cancer Hospital.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Sun K, Zheng R, et al. Cancer incidence and mortality in China, 2014. Chin J Cancer Res 2018;30:1-12. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Ruttkay-Nedecky B, Jimenez Jimenez AM, Nejdl L, et al. Relevance of infection with human papillomavirus: the role of the p53 tumor suppressor protein and E6/E7 zinc finger proteins Int J Oncol 2013;43:1754-62. (Review). [Crossref] [PubMed]
- Marcel V, Nguyen Van Long F, Diaz JJ. 40 Years of Research Put p53 in Translation. Cancers (Basel) 2018;10. [Crossref] [PubMed]
- Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell 2017;170:1062-78. [Crossref] [PubMed]
- Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature 1979;278:261-3. [Crossref] [PubMed]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2010;2:a001008. [Crossref] [PubMed]
- Molina-Vila MA, Bertran-Alamillo J, Gasco A, et al. Nondisruptive p53 Mutations Are Associated with Shorter Survival in Patients with Advanced Non-Small Cell Lung Cancer. Clin Cancer Res 2014;20:4647-59. [Crossref] [PubMed]
- Balaburski GM, Hontz RD, Murphy ME. p53 and ARF: unexpected players in autophagy. Trends Cell Biol 2010;20:363-9. [Crossref] [PubMed]
- Wang Q, Zou Y, Nowotschin S, et al. The p53 Family Coordinates Wnt and Nodal Inputs in Mesendodermal Differentiation of Embryonic Stem Cells. Cell Stem Cell 2017;20:70-86. [Crossref] [PubMed]
- Liu J, Zhang C, Hu W, et al. Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Lett 2015;356:197-203. [Crossref] [PubMed]
- Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol 2013;15:2-8. [Crossref] [PubMed]
- Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev 2012;26:1268-86. [Crossref] [PubMed]
- Yue X, Zhao Y, Xu Y, et al. Mutant p53 in Cancer: Accumulation, Gain-of-Function, and Therapy. J Mol Biol 2017;429:1595-606. [Crossref] [PubMed]
- Blandino G, Deppert W, Hainaut P, et al. Mutant p53 protein, master regulator of human malignancies: a report on the Fifth Mutant p53 Workshop. Cell Death Differ 2012;19:180-3. [Crossref] [PubMed]
- Dobes P, Podhorec J, Coufal O, et al. Influence of mutation type on prognostic and predictive values of TP53 status in primary breast cancer patients. Oncol Rep 2014;32:1695-702. [Crossref] [PubMed]
- Uehara I, Tanaka N. Role of p53 in the Regulation of the Inflammatory Tumor Microenvironment and Tumor Suppression. Cancers 2018;10. [Crossref] [PubMed]
- Poeta ML, Manola J, Goldwasser MA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med 2007;357:2552-61. [Crossref] [PubMed]
- Skinner HD, Sandulache VC, Ow TJ, et al. TP53 disruptive mutations lead to head and neck cancer treatment failure through inhibition of radiation-induced senescence. Clin Cancer Res 2012;18:290-300. [Crossref] [PubMed]
- Malekzadeh P, Pasetto A, Robbins PF, et al. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J Clin Invest 2019;129:1109-14. [Crossref] [PubMed]
- Zhao Z, Wang P, Gao Y, et al. The high expression instead of mutation of p53 is predictive of overall survival in patients with esophageal squamous-cell carcinoma: a meta-analysis. Cancer Med 2017;6:54-66. [Crossref] [PubMed]
- Su D, Zhang D, Chen K, et al. High performance of targeted next generation sequencing on variance detection in clinical tumor specimens in comparison with current conventional methods. J Exp Clin Cancer Res 2017;36:121. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- Zhu J, Sammons MA, Donahue G, et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 2015;525:206-11. [Crossref] [PubMed]
- Zhang F, Ying L, Jin J, et al. GAP43, a novel metastasis promoter in non-small cell lung cancer. J Transl Med 2018;16:310. [Crossref] [PubMed]
- Taylor NJ, Nikolaishvili-Feinberg N, Midkiff BR, et al. Rational manual and automated scoring thresholds for the immunohistochemical detection of TP53 missense mutations in human breast carcinomas. Appl Immunohistochem Mol Morphol 2016;24:398-404. [Crossref] [PubMed]
- Huang K, Chen L, Zhang J, et al. Elevated p53 expression levels correlate with tumor progression and poor prognosis in patients exhibiting esophageal squamous cell carcinoma. Oncol Lett 2014;8:1441-6. [Crossref] [PubMed]
- John T, Liu G, Tsao MS. Overview of molecular testing in non-small-cell lung cancer: mutational analysis, gene copy number, protein expression and other biomarkers of EGFR for the prediction of response to tyrosine kinase inhibitors. Oncogene 2009;28 Suppl 1:S14-23. [Crossref] [PubMed]
- Elsensohn A, Shiu J, Grove N, et al. Distinguishing Neurofibroma From Desmoplastic Melanoma: The Value of p53. Am J Surg Pathol 2018;42:372-5. [Crossref] [PubMed]
- Hirsch FR, Varella-Garcia M, Bunn PA Jr, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol 2003;21:3798-807. [Crossref] [PubMed]
- Youden WJ. Index for rating diagnostic tests. Cancer 1950;3:32-5. [Crossref] [PubMed]
- Soussi T, Legros Y, Lubin R, et al. Multifactorial analysis of p53 alteration in human cancer: a review. Int J Cancer 1994;57:1-9. [Crossref] [PubMed]
- Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res 2000;77:81-137. [Crossref] [PubMed]
- Sengpiel C, König IR, Rades D, et al. p53 Mutations in Carcinoma of the Esophagus and Gastroesophageal Junction. Cancer Invest 2009;27:96-104. [Crossref] [PubMed]
- Gao YB, Chen ZL, Li JG, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 2014;46:1097-102. [Crossref] [PubMed]
- Maeng CH, Lee J, van Hummelen P, et al. High-Throughput Genotyping in Metastatic Esophageal Squamous Cell Carcinoma Identifies Phosphoinositide-3-Kinase and BRAF Mutations. PLoS One 2012;7:e41655. [Crossref] [PubMed]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009;9:701-13. [Crossref] [PubMed]
- Kang N, Wang Y, Guo S, et al. Mutant TP53 G245C and R273H promote cellular malignancy in esophageal squamous cell carcinoma. BMC Cell Biol 2018;19:16. [Crossref] [PubMed]
- Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009;9:749-58. [Crossref] [PubMed]
- Wu XC, Zheng YF, Tang M, et al. Association Between Smoking and p53 Mutation in Oesophageal Squamous Cell Carcinoma: A Meta-analysis. Clin Oncol (R Coll Radiol) 2015;27:337-44. [Crossref] [PubMed]
- Canale M, Petracci E, Delmonte A, et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin Cancer Res 2017;23:2195-202. [Crossref] [PubMed]
- Fisher OM, Lord SJ, Falkenback D, et al. The prognostic value of TP53 mutations in oesophageal adenocarcinoma: a systematic review and meta-analysis. Gut 2017;66:399-410. [Crossref] [PubMed]
- Végran F, Rebucci M, Chevrier S, et al. Only Missense Mutations Affecting the DNA Binding Domain of P53 Influence Outcomes in Patients with Breast Carcinoma. PLoS One 2013;8:e55103. [Crossref] [PubMed]
- Trbusek M, Smardova J, Malcikova J, et al. Missense mutations located in structural p53 DNA-binding motifs are associated with extremely poor survival in chronic lymphocytic leukemia. J Clin Oncol 2011;29:2703-8. [Crossref] [PubMed]
- Bueno AM, Molina M, Fielding A, et al. LBA42Disruptive mutations in TP53 associate with survival benefit in a PARPi trial in ovarian cancer. Ann Oncol 2017;28:mdx440.036.
- Takami H, Yoshida A, Fukushima S, et al. Revisiting TP53 Mutations and Immunohistochemistry--A Comparative Study in 157 Diffuse Gliomas. Brain Pathol 2015;25:256-65. [Crossref] [PubMed]
- Olivier M, Taniere P. Somatic mutations in cancer prognosis and prediction: lessons from TP53 and EGFR genes. Curr Opin Oncol 2011;23:88-92. [Crossref] [PubMed]


