
NUCB2 polymorphisms are associated with an increased risk for type 2 diabetes in the Chinese population
Introduction
Type 2 diabetes mellitus (T2DM) is a prevalent complex metabolic disorder that is triggered by the combination of strong genetic predisposition and environmental factors. Notably, numerous genetic association studies have been conducted, and more than 200 genetic loci for T2DM identified within the last decade (1). However, these studies have not fully explained the heritability of T2DM, and this subject remains to be further elucidated.
The nucleobindin 2 (NUCB2) is a polypeptide composed of 396 amino acid and is a precursor of nesfatin-1, nesfatin-2 and nesfatin-3, spanning residues 1–82, 85–163 and 85–163, respectively (2). An initial study proposed that among the three peptides, only nesfatin-1 is a physiologic modulator of satiety. It was shown that intracerebroventricular injection of rats with nesfatin-1 suppresses food intake, whereas injection with nesfatin-1 neutralizing antibody significantly stimulates appetite (2). Subsequently, the anorexigenic effects of nesfatin-1 were confirmed in further studies (3-6). Nevertheless, Ravussin et al. revealed that loss or knockdown of NUCB2 in orexigenic neurons does not affect food intake or adiposity, whereas the loss of NUCB2 in myeloid cells mediates insulin resistance in response to high-fat diet (7). In addition to, the central nervous system (CNS), NUCB2 mRNA was detected in peripheral tissues, including the adipocytes, gastric endocrine cells, and islet cells (8,9). Notably, NUCB2 is selectively expressed in pancreatic islet beta cells (10,11) and may play a role in glucose homeostasis, as has been shown in in vivo and in in vitro studies. Using mouse isolated islets and MIN6 cells, Nakata et al. (12) showed that nesfatin-1 significantly enhanced glucose-stimulated insulin secretion in vitro. The induction mechanism could be related to promoting Ca2+ influx through L-type Ca2+ channels independent of protein kinase A (PKA) and phospholipase A2 (PLA2) in mice islet beta cells. Moreover, intravenous injection with nesfatin-1 significantly reduces blood glucose in hyperglycemic db/db mice (13). A recent study showed that beta-cell-specific NUCB2 knockout mice exhibited late-onset elevation of casual blood glucose, elevated blood glucose and lowered insulin secretion in glucose tolerance test (14). Besides, intracellular NUCB2 mRNA levels and protein synthesis or release in the pancreatic beta cells are dynamically regulated by glucose levels. In vitro, the expression of human islet NUCB2 mRNA is upregulated under gluco-lipotoxic conditions, and is down-regulated in T2DM patients compared to controls (11). Similarly, mRNA and protein expression is significantly reduced in the islets of Goto-Kakizaki (GK) rats, a model of type 2 diabetes (8). It has also been shown that fasting plasma nesfatin-1 levels are significantly reduced in T2DM (15-17), whereas plasma nesfatin-1 in type 1 diabetic patients was unchanged compared with healthy individuals (17). Accordingly, the concentrations of nesfatin-1 in the serum and milk of gestational diabetic lactating women is lower than that of control participants (18). It was shown that nesfatin-1 could be a potential novel biomarker for the prediction and early diagnosis of gestational diabetes mellitus (19). As revealed in the findings of a systematic review and meta-analysis, there is a relationship between circulating nesfatin-1 levels and type 2 diabetes (20). The evidence presented in this section suggests the possible role of nesfatin-1 in the pathogenesis of T2DM.
Zegers et al. (21) explored the effects of NUCB2 polymorphisms on 1049 obese Caucasian patients and 315 normal-weight controls. The study found an association between obesity and three NUCB2 single nucleotide polymorphisms (SNPs) (rs1330, rs214101, and rs757081), only when data from the male subpopulation was analyzed separately. A recent study demonstrated the correlation between the rs757081 variant of the NUCB2 gene and childhood adiposity in a cohort of severely obese Chinese children in Singapore and non-obese Chinese children from Da Qing, China (22). Besides, Wang et al. (23) found that rs757081 in NUCB2 was associated with the risk for developing T2DM in the Chinese Han population. Consequently, the present study hypothesized that NUCB2 polymorphisms could be associated with the risk for T2DM. We used a cohort of 578 T2DM patients and 1,609 healthy controls to evaluate the contribution of NUCB2 polymorphisms to the development of T2DM.
Methods
Study subjects
A case-control study approach was adopted to obtain an in-depth understanding of the heritability of T2DM. Our sample population consisted of 578 T2DM patients and 1,609 non-diabetic healthy controls. None of the participants were genetically related to each other. The primary diagnostic criteria for patients with T2DM were either a fasting plasma glucose level ≥7.0 mmol/L or a 2-h postprandial plasma glucose level ≥11.1 mmol/L. The study also recruited patients who had a clinical history of T2DM and were on anti-diabetic treatments such as oral hypoglycemic treatments or parental insulin. Patients with type 1 diabetes mellitus were, however, excluded. All participants were Chinese Han population. Before undertaking the study, ethical approval was obtained from the hospital Ethics Board and written informed consent was obtained from each patient. A detailed description of the study and control population characteristics is presented in Table 1.

Full table
Anthropometric and clinical measurements
Anthropometric measurements, including height and weight, were taken using standard procedures. The body mass index (BMI) was calculated using the formula: BMI = kg/m2, where Kg is the weight in kilograms, while m2 is a square of the height in meters. Plasma samples were collected following a 12-h overnight fast, and glucose levels were assessed on an automatic enzymatic analyzer. The glycohemoglobin (HbA1c) test was done following a standard procedure.
SNP selection, genotyping, and quality control (QC) filters
We selected SNPs according to the following procedures. First, NUCB2 SNPs and genotypes were identified from the HapMap-HCB. Second, the linkage disequilibrium (LD) between SNPs was calculated using Genome Variation Server 138, and all of the monomorphic sites were filtered. A null allele frequency >5% and a standard LD threshold of r2 >0.80 were used to select five candidate tag SNPs (rs10832756, rs1330, rs10766383, rs10832757, and rs11024251) based on previous genetic studies (21,24).
Subsequently, the following QC filters were applied to eliminate low quality SNPs and samples before analysis. The same QC parameters were applied to both scans and excluded SNPs with (I) a missing call rate ≥2%; (II) >1 discordance; (III) significant deviations from the Hardy-Weinberg equilibrium (HWE) (P<0.001); (IV) a minor allele frequency <1%. All SNPs (rs10832756, rs1330, rs10766383, rs10832757, and rs11024251) passed the QC criteria and were included in the analysis.
The concentration and quality of genomic DNA extracted from whole blood were assessed by resolution on a 1.5% agarose gel. SNP genotyping was performed on the MassARRAY system, as described by Elis et al. (25).
Statistical analysis
Baseline clinical data are presented as mean ± standard deviation (SD). Non-normally distributed data were subjected to a log transformation before statistical testing. The value differences between the T2DM and control groups were compared using an independent samples t-test or chi-squared test. For each SNP, HWE was calculated in the control group; all SNPs were in HWE. Independent risk factors for developing T2DM were identified by logistic regression analysis. Linear regression analysis was used to determine whether the variants influenced BMI. Statistical was performed by the SPSS software version 22.0 (IBM Corp., Armonk, NY, USA), and a P value <0.05 was considered statistically significant. LD was assessed by calculating r2 using Haploview v.4.2 (Broad Institute, Cambridge, MA, USA). The Quanto software version 1.2.4 was used for power calculations in genome-wide association studies (GWAS). Association analysis for the genotypes of the five identified SNPs was performed using the PLINK software v.1.07 (26).
Results
Baseline clinical characteristics
The anthropometric and biochemical characteristics of the participants are detailed in Table 1. Statistically significant (P<0.05) differences in the age, gender, and BMI of participants in the two groups were noted. Thus, all P values were age- or gender-adjusted before analysis.
Characteristics of the SNPs
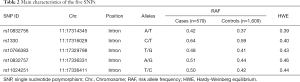
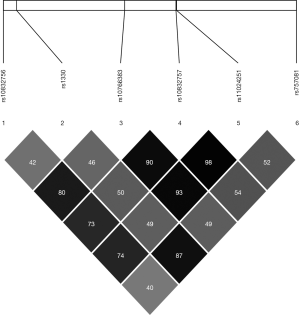
The position, risk allele frequency (RAF), and HWE characteristics of the SNPs are presented in Table 2. Genotype distribution in both the case and control groups conformed to HWE. The LD patterns of the SNPs were assessed in the sample populations using r2 values (Figure 1). The SNP rs1330 was in weak LD with all the other four SNPs (rs10832756, rs10766383, rs10832757, and rs11024251). SNP rs1330 was in high linkage with rs757081(r2 >0.8), which is the only SNP previously reported to be linked to diabetes in Chinese Han populations.
Full table
Association of 5 SNPs with T2DM
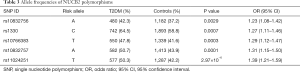
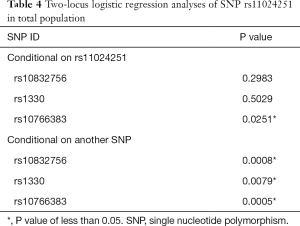
The allele frequencies of NUCB2 polymorphisms are shown in Table 3. Our results of Fisher’s exact test indicate that SNPs rs10832756 [odds ratio (OR) 1.23, 95% confidence interval (CI) 1.08–1.42, P=0.0029], rs1330 (OR 1.27, 95% CI: 1.11–1.46, P=0.0007), rs10766383 (OR 1.29, 95% CI: 1.12–1.47, P=0.0003), rs10832757 (OR 1.31, 95% CI: 1.15–1.50, P=0.0001), and rs11024251 (OR 1.39, 95% CI: 1.21–1.59, P=2.97×10−6) are associated with an increased risk of developing T2DM (Table 3). We analyzed the 5 SNPs via two-locus logistic regression analyses. The SNP rs11024251 was entered individually into the regression model as the best NUCB2 marker, and all other markers were sequentially added to assess whether a second locus could improve the model. Further, we tested the regression model as conditional on each of the five loci individually and added the test locus. As shown in Table 4, all the markers could be improved by adding SNP rs11024251. The genotype distributions of the five SNPs in the control group and T2DM patients are shown in Table 5. We found that the AA genotypes of rs10832756 and rs10832757, the CC genotype of rs1330, and the TT genotypes of rs10766383 and rs11024251 were associated with an increased risk for diabetes (P<0.05). These findings suggest that both allele frequencies and genotype distributions of NUCB2 polymorphisms are significantly different between the two groups.
Full table
Full table

Full table
Sex dimorphism in the associations of 5 SNPs with T2DM
Our logistic regression analysis (adjusted for age and gender) results showed that all the five SNPs are associated with T2DM (Table 6). Analysis of the data for males and females separately, however, showed that only two SNPs (rs10832757 and rs11024251) are associated with T2DM in the male population (361 cases vs. 398 controls) and four SNPs (rs1330, rs10766383, rs10832757, and rs11024251) are associated with T2DM in the female population (217 cases vs. 1,211 controls; Table 6). These findings, therefore, suggest that the NUCB2 SNPs related to the risk for T2DM are different between males and females.
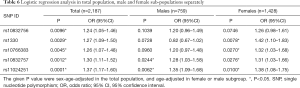
Full table
Linear regression analysis on BMI
This study further developed a linear regression model to evaluate the effect of the NUCB2 variants on T2DM-related parameters. Although our results showed that different genotypes of the five SNPs are not significantly associated with BMI, we found that the SNP rs1330 is significantly associated with BMI in female patients (Table 7).

Full table
Discussion
This case-control study of 578 T2DM patients and 1,609 healthy controls involved five NUCB2 SNPs and identified an association between polymorphisms in the NUCB2 gene and an increased risk for type 2 diabetes in Chinese populations. All the five SNPs were associated with an increased risk of developing T2DM at different statistical differences. The SNP rs11024251 showed the highest susceptibility to T2DM since it improved models with any of the other SNPs and had the lowest P-value among all the NUCB2 SNPs. Notably, our logistic and linear regression analyses result for the entire population, and male and female subpopulations separately suggested that NUCB2 SNPs associated with the risk for T2DM have gender disparity.
Nesfatin-1, an 82-amino acid peptide derived from the post-translational processing of NUCB2, was initially reported by Oh-I et al. in 2006 (2). Accumulating evidence has revealed that NUCB2 is not only a novel satiety factor widely expressed in the CNS but is also expressed in peripheral tissues where it regulates glucose and energy metabolism. Recently, it has been reported that infusion of nesfatin-1 into the third cerebral ventricle markedly promotes muscle glucose uptake, inhibits hepatic glucose production, and inhibits hepatic phosphoenolpyruvate carboxykinase (PEPCK) mRNA and protein. This infusion also inhibits enzymatic activity of PEPCK in control and diet-induced obese rats (27). Another study showed that insulin and high glucose levels activate paraventricular nucleus (PVN) nesfatin-1 and proopiomelanocortin (POMC) neurons (28), suggesting that nesfatin-1 neurons cooperate with melanocortin neurons in the regulation of glucose metabolism. Besides the extensive distribution in the CNS, NUCB2 is also expressed in the periphery, including the stomach, pancreas, testis, and adipose tissues (8,9,29). Indeed, NUCB2 is colocalized almost exclusively with insulin in the beta-cells of pancreatic islets (8,10,11). A recent study showed that beta-cell-specific NUCB2 knockout mice have elevated blood glucose levels and reduced insulin secretion (14). Also, diabetic GK rats have lower NUCB2 protein levels in pancreatic islets compared to non-diabetic controls (8). Besides, plasma nesfatin-1 concentrations and islet NUCB2 mRNA are significantly decreased in T2DM patients compared with healthy controls (11,17,30). These previous findings indicate that NUCB2 plays a significant role in the development of T2DM. Similarly, our results indicated that the five tag SNPs of NUCB2 are associated with an increased risk for T2DM. Thus, dysfunction of expression, secretion, and/or action of NUCB2 might be involved in the development and progression of T2DM.
The findings of previous studies report that NUCB2 polymorphisms are associated with obesity in adults and children (21,22). It has been shown that SNPs rs1330, rs214101, and rs757081 are associated with BMI, weight, and fat-free mass in Caucasian males (21). The c.1012C>G polymorphism of the NUCB2 gene (rs757081) is correlated with childhood adiposity (22). However, the correlation between NUCB2 polymorphisms and the pathogenesis of T2DM is obscure. Recently, a study indicated that the NUCB2 SNP rs757081 is associated with a reduced risk of developing T2DM in a Chinese Han population (23), which provides evidence of the relationship between NUCB2 polymorphisms and T2DM.
Elucidation of the genetic pathways that influence the risk to T2DM could provide a better understanding of the pathophysiology of the disease and identify possible pharmacologic targets for its treatment. Thus, we explored the association between NUCB2 polymorphisms and the development of T2DM in a Chinese Han population. Based on reports from previous genetic studies (21,24), we investigated five candidate tag SNPs (rs10832756, rs1330, rs10766383, rs10832757, and rs11024251) of NUCB2.
First, we showed that 5 SNPs had significantly different allele frequencies (at a P=0.05) in T2DM patients and healthy controls; the SNP rs11024251 showed the strongest association. These results led us to further test a regression model composed of one SNP, then added each one of the remaining four SNPs in turn, then adding the testing locus. We found that the SNP rs11024251, as the best marker for NUCB2, improved the other SNPs in the regression model. Our findings indicate that the five NUCB2 SNPs are associated with an increased risk for T2DM. We, however, did not rule out the possibility that NUCB2 variants could be related to diabetes through effects on BMI because the BMI was significantly different between the control and diabetic groups.
When the data for male and female subpopulations were separately analyzed, we identified two loci (including the SNP, rs1330 and the SNP, rs10766383) whose associations with T2DM were only evident in the female subgroup but were not significant (P>0.05) in males. In addition, we only found significant P values for the SNP rs1330, in association with the BMI in females. Similarly, a study by Zegers et al. in 2011 (21) observed a gender disparity in the NUCB2 polymorphisms associated with obesity.
At present, it is not clear why we observed different significant SNPs between different gender subgroups or why we only found an association between females and BMI. This observation could be explained by the different sex hormone levels in different genders. A previous study revealed a sex-specific regulation of nesfatin-1 with higher levels in females (31). An ex vivo experiment showed that the expression of NUCB2 mRNA in cultured pituitary glands is reduced by increasing concentrations of progesterone in culture media, but the opposite effect occurred with 17b-estradiol (32). The testosterone changes were also associated with the initiation of puberty regulated by the production of NUCB2/nesfatin-1 via adipose tissue and NUCB2 gastric output (33). Another possible explanation for this gender disparity could be that variations in NUCB2 could have sex-specific phenotypic effects in T2DM, as illuminated in several other diseases (34,35). But the hypothesis should be verified via large sample size studies in the future. Together, these data suggest that reproductive-endocrine regulation may contribute to sex-specific regulation of NUCB2. However, the specific underlying mechanism remains to be established.
The findings presented in this study could somewhat be limited by the following. First, our study was conducted in a Chinese Han population, and caution should be exercised when extrapolating the data to other communities. Second, our sample size was relatively small, which may mask significant differences. Therefore, a higher sample size could provide more information in future studies. Our study did not explore all the concerns around the current subject, and further research should, therefore, determine whether any observed differences in genetic associations are influenced by sex in Chinese Han populations.
Conclusions
In the current study, we investigated whether the risk of T2DM in the Chinese Han population is linked to the five SNPs of NUCB2. We found that these SNPs of NUCB2 play a significant role in the risk for T2DM, especially the SNP, rs11024251. Besides, we found the associated SNPs were different between males and females. Moreover, further research should explore the pathophysiological regulation of NUCB2 in the presence of T2DM. Future studies also need to determine the sex-specific influence of NUCB2 on the risk for T2DM, which might contribute to precise prevention and individual-tailored treatment for T2DM.
Acknowledgements
We thank all patients and normal individuals for participating in this study.
Funding: This work was supported in part by Shanghai Science and Technology Committee (18dz2304400, 114119b0400, 15411970700 to JL Yang); the National Natural Science Foundation of China (81870537, 31571296 to SX Zhao); and Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20161318 to SX Zhao).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Our study was approved by the Ethics Committee of Shanghai Ruijin Hospital Group Minhang District Central Hospital (No. 20150211) and written informed consent was obtained from each patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 2018;50:1505-13. [Crossref] [PubMed]
- Oh-I S, Shimizu H, Satoh T, et al. Identification of nesfatin-1 as a satiety molecule in the hypothalamus. Nature 2006;443:709-12. [Crossref] [PubMed]
- Könczöl K, Pintér O, Ferenczi S, et al. Nesfatin-1 exerts long-term effect on food intake and body temperature. Int J Obes (Lond) 2012;36:1514-21. [Crossref] [PubMed]
- Gotoh K, Masaki T, Chiba S, et al. Nesfatin-1, corticotropin-releasing hormone, thyrotropin-releasing hormone, and neuronal histamine interact in the hypothalamus to regulate feeding behavior. J Neurochem 2013;124:90-9. [Crossref] [PubMed]
- Wernecke K, Lamprecht I, Jöhren O, et al. Nesfatin-1 increases energy expenditure and reduces food intake in rats. Obesity 2014;22:1662-8. [Crossref] [PubMed]
- Nakata M, Gantulga D, Santoso P, et al. Paraventricular NUCB2/Nesfatin-1 Supports Oxytocin and Vasopressin Neurons to Control Feeding Behavior and Fluid Balance in Male Mice. Endocrinology 2016;157:2322-32. [Crossref] [PubMed]
- Ravussin A, Youm YH, Sander J, et al. Loss of Nucleobindin-2 Causes Insulin Resistance in Obesity without Impacting Satiety or Adiposity. Cell Rep 2018;24:1085-92. [PubMed]
- Foo KS, Brauner H, Ostenson CG, et al. Nucleobindin-2/nesfatin in the endocrine pancreas: distribution and relationship to glycaemic state. J Endocrinol 2010;204:255-63. [Crossref] [PubMed]
- Tsuchiya T, Shimizu H, Yamada M, et al. Fasting concentrations of nesfatin-1 are negatively correlated with body mass index in non-obese males. Clin Endocrinol 2010;73:484-90. [PubMed]
- Gonzalez R, Tiwari A, Unniappan S. Pancreatic beta cells colocalize insulin and pronesfatin immunoreactivity in rodents. Biochem Biophys Res Commun 2009;381:643-8. [Crossref] [PubMed]
- Riva M, Nitert MD, Voss U, et al. Nesfatin-1 stimulates glucagon and insulin secretion and beta cell NUCB2 is reduced in human type 2 diabetic subjects. Cell Tissue Res 2011;346:393-405. [Crossref] [PubMed]
- Nakata M, Manaka K, Yamamoto S, et al. Nesfatin-1 enhances glucose-induced insulin secretion by promoting Ca(2+) influx through L-type channels in mouse islet β-cells. Endocr J 2011;58:305-13. [Crossref] [PubMed]
- Aydin S. Multi-functional peptide hormone NUCB2/nesfatin-1. Endocrine 2013;44:312-25. [Crossref] [PubMed]
- Yang Y, Zhang B, Nakata M, et al. Islet β-cell-produced NUCB2/nesfatin-1 maintains insulin secretion and glycemia along with suppressing UCP-2 in β-cells. J Physiol Sci 2019;69:733-9. [Crossref] [PubMed]
- Algul S, Ozkan Y, Ozcelik O. Serum nesfatin-1 levels in patients with different glucose tolerance levels. Physiol Res 2016;65:979-85. [Crossref] [PubMed]
- Dai R, Deng G, Sun Z, et al. Relation of serum and vitreous nesfatin-1 concentrations with diabetic retinopathy. J Clin Lab Anal 2017;31. [PubMed]
- Li QC, Wang HY, Chen X, et al. Fasting plasma levels of nesfatin-1 in patients with type 1 and type 2 diabetes mellitus and the nutrient-related fluctuation of nesfatin-1 level in normal humans. Regul Pept 2010;159:72-7. [Crossref] [PubMed]
- Aydin S. The presence of the peptides apelin, ghrelin and nesfatin-1 in the human breast milk, and the lowering of their levels in patients with gestational diabetes mellitus. Peptides 2010;31:2236-40. [Crossref] [PubMed]
- Mierzyński R, Poniedziałek-Czajkowska E, Dłuski D, et al. Nesfatin-1 and Vaspin as Potential Novel Biomarkers for the Prediction and Early Diagnosis of Gestational Diabetes Mellitus. Inter J Mol Sci 2019. doi:
10.3390/ijms20010159 . - Zhai T, Li SZ, Fan XT, et al. Circulating Nesfatin-1 Levels and Type 2 Diabetes: A Systematic Review and Meta-Analysis. J Diabetes Res 2017;2017:7687098.
- Zegers D, Beckers S, Mertens IL, et al. Association between polymorphisms of the Nesfatin gene, NUCB2, and obesity in men. Mol Genet Metab 2011;103:282-6. [Crossref] [PubMed]
- Chen YY, Chan RME, Tan KML, et al. The association of a nucleobindin 2 gene (NUCB2) variant with childhood adiposity. Gene 2013;516:48-52. [Crossref] [PubMed]
- Wang C, Wang Y, Hu W. Association of the polymorphism in NUCB2 gene and the risk of type 2 diabetes. Diabetol Metab Syndr 2017;9:39. [Crossref] [PubMed]
- Du H, Vimaleswaran KS, Angquist L, et al. Genetic polymorphisms in the hypothalamic pathway in relation to subsequent weight change--the DiOGenes study. PLoS One 2011;6:e17436. [Crossref] [PubMed]
- Ellis JA, Ong B. The MassARRAY System for Targeted SNP Genotyping. Methods Mol Biol 2017;1492:77-94. [Crossref] [PubMed]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559-75. [Crossref] [PubMed]
- Yang M, Zhang Z, Wang C, et al. Nesfatin-1 action in the brain increases insulin sensitivity through Akt/AMPK/TORC2 pathway in diet-induced insulin resistance. Diabetes 2012;61:1959-68. [Crossref] [PubMed]
- Gantulga D, Maejima Y, Nakata M, et al. Glucose and insulin induce Ca2+ signaling in nesfatin-1 neurons in the hypothalamic paraventricular nucleus. Biochem Biophys Res Commun 2012;420:811-5. [Crossref] [PubMed]
- Kim J, Chung Y, Kim H, et al. The Tissue Distribution of Nesfatin-1/NUCB2 in Mouse. Dev Reprod 2014;18:301-9. [Crossref] [PubMed]
- Liu F, Yang Q, Gao N, et al. Decreased plasma nesfatin-1 level is related to the thyroid dysfunction in patients with type 2 diabetes mellitus. J Diabetes Res 2014;2014:128014.
- Hofmann T, Elbelt U, Ahnis A, et al. Sex-specific regulation of NUCB2/nesfatin-1: Differential implication in anxiety in obese men and women. Psychoneuroendocrinology 2015;60:130-7. [Crossref] [PubMed]
- Chung Y, Kim J, Im E, et al. Progesterone and 17β-estradiol regulate expression of nesfatin-1/NUCB2 in mouse pituitary gland. Peptides 2015;63:4-9. [Crossref] [PubMed]
- Senin LL, Al-Massadi O, Barja-Fernandez S, et al. Regulation of NUCB2/nesfatin-1 production in rat's stomach and adipose tissue is dependent on age, testosterone levels and lactating status. Mol Cell Endocrinol 2015;411:105-12. [Crossref] [PubMed]
- Ng SK, Burdon KP, Fitzgerald JT, et al. Genetic Association at the 9p21 Glaucoma Locus Contributes to Sex Bias in Normal-Tension Glaucoma. Invest Ophthalmol Vis Sci 2016;57:3416-21. [Crossref] [PubMed]
- Gautam Y, Afanador Y, Abebe T, et al. Genome-wide analysis revealed sex-specific gene expression in asthmatics. Hum Mol Genet 2019; [Epub ahead of print]. [Crossref] [PubMed]


