
Downregulation of ubiquitin-specific protease 2 possesses prognostic and diagnostic value and promotes the clear cell renal cell carcinoma progression
Introduction
Each year, more than 400,000 new cancer diagnoses globally, accounting for more than 175,000 deaths, were afflicted by renal cell carcinoma (RCC) (1,2). Among all RCC subtypes, the clear cell renal cell carcinoma (ccRCC) accounts for the highest proportion (approximately 70–80%), mortality rate, invasion, and metastasis (3). Moreover, reports indicate that approximately 30% of ccRCC patients still develop recurrence after surgical treatment (4). It is, therefore, important to discover more effective molecular biomarkers to aid early detection and diagnosis, monitor metastasis and recurrence, and promote postoperative adjuvant targeted therapy.
Deubiquitination, a post-translational modification, is known to highly regulate mechanisms of cell progression by inhibiting ubiquitination and degradation of protein substrates (5). To date, several deubiquitinating enzymes (DUBs) have been implicated in human benign and malignant diseases, and their substrates and functions have been extensively studied over the past couple of decades (6). Ubiquitin-specific protease 2 (USP2), initially detected in rat testis (7), has been reported to regulate various cellular events, including cell-cycle and proliferation (8), DNA repair (9), modulation of the circadian rhythm (10-13), inflammatory responses (14-16), regulation of lipoprotein clearance (17), as well as maintenance of normal sodium balance and blood pressure (18). Moreover, USP2 has shown oncogenic properties in many cancer types, particularly breast cancer and prostate cancer, by stabilizing fatty acid synthase (19), EGFR (5), MDM2 (20) and MDM4 (21). However, it has been shown that ectopic expression of USP2a in the breast cancer cell line, MCF7, inhibited the NF-κB signaling pathway and thus exhibited a unique anticancer characteristic (22,23). These results suggest that USP2 has a more complicated function in human carcinoma, beyond carcinogenesis and anti-apoptotic effects (21). Despite accumulating evidence on the role of USP2 in several types of cancer, its underlying effects and specific mechanisms in ccRCC remain uncharacterized. Previous studies have shown that the different functions of USP2 may depend on the cellular context (24), emphasizing the necessity for further exploration into its function in ccRCC.
In this study, we focused on the role of USP2 in tumor development biology in human ccRCC. Our results revealed that USP2 was downregulated in ccRCC tissues and renal cancer cell lines. Moreover, we identified low-expression of USP2 possesses crucial prognostic and diagnostic value in ccRCC. Furthermore, USP2 acted as an anti-oncogene in ccRCC, with its overexpression inhibiting proliferation, migration, and invasion of ccRCC cells.
Methods
Tissue samples
A total of 30 pairs of clinical renal cancer samples, collected between 2017–2018, were obtained from the Department of Urology, Union Hospital, Tongji Medical College (Wuhan, China). The resected samples were divided into two parts. The first set of samples was immediately frozen in liquid nitrogen for use in RNA extraction and Western blotting experiments. The second set was fixed in formalin and embedded in paraffin, then used for immunohistochemistry assays. No adjuvant anticancer therapy was received before surgery. Prior to the study, every patient’s fully informed consent was obtained, and the study approval by the Institutional Review Board of Huazhong University of Science and Technology.
Immunohistochemical staining assays
The second batch of sampled tissues, described above, was incubated with rabbit USP2 polyclonal antibody (ABclonal, A10399, 1:1,000) overnight at 4 °C, tissue sections washed with PBS then incubated with anti-rabbit secondary antibodies at room temperature for 2 hours. Color change was monitored by 3,3-N-Diaminobenzidine Tertrahydrochloride (DAB), with samples showing a dark brown coloration regarded as positive.
Cell culture
A normal (HK2) and ccRCC cell lines (ACHN, 786-O, CAKi-1, and A498) were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in high-glucose DMEM. The high-glucose DMEM was supplemented with 10% FBS and 1% penicillin-streptomycin into. All cells were cultured in an incubator at 37 °C, and 5% CO2.
Transient transfection assays
We cultured Caki-1 and A498 cells in 6-well plates, at approximately 60–70% confluence, then transfected them with 4 µg of expression plasmids harboring USP2 (Genechem, China), using Lipofectamine 2000 (Invitrogen, CA, USA). Cells were collected 48 hours after transfection. Subsequent assays of total RNA and protein expression levels were performed by western blot and qRT-PCR.
RNA Isolation and qRT-PCR
Total RNA was first isolated from cell lines and tissues using the TRizol reagent (Thermo; Massachusetts, USA), then 2 µg of the RNA reverse transcribed to cDNA. The SYBR Green mix (Thermo, Massachusetts, USA) was used for quantitative real-time polymerase chain reaction (qRT-PCR) analysis of USP2, with GAPDH included as an internal amplification control. Oligonucleotide primers used in the analysis were purchased from Tianyi Huiyuan, and are listed as follows:
GAPDH: left primer 5'-CCAGAACAGCATCCCTGCCT-3'; right primer 5'-CCTGCTTCACCACCTTCTTG-3';
USP2: left primer 5'-CCGCGCTTTGTTGGCTATAA-3'; right primer 5'-CCCGATCCTACTGTCTTCCC-3'.
Western blot analysis
Firstly, cell and tissue proteins were pyrolysed in RIPA protein lysis buffer (Beyotime Institute of Biotechnology; Haimen, China) in a mixture containing 1 mM protease inhibitor cocktail and PMSF. Secondly, 30 µg of protein per hole was subjected to 10% SDS-PAGE gel electrophoresis then transferred onto PVDF membranes at 250 mA for 90 min. The membranes were blocked in PBST, containing 5% nonfat milk, at room temperature and after 2 hours, they were incubated with primary antibodies against USP2 (ABclonal, A10399, 1:1,000), GAPDH (Proteintech, 60004-1-Ig, 1:1,000) at 4 °C for at least 12 hours. Finally, they were washed three times, and then incubated with a secondary antibody for 2 hours before detection by ChemiDoc-XRS+ (Bio-Rad, USA).
Cell proliferation assays
Briefly, 2000 cells were added to each hole in the 96-well plates, 48 hours after transfection. Cell proliferation capacity was then assessed using the CCK-8 assay at 0, 24, 48, 72 and 96 hours following treatments, respectively, according to the manufacturer’s instructions.
Migration and invasion assays
These assays were performed on cells 48 hours after transfection and 24 hours after starvation. Briefly, cells were resuspended with serum-free medium, then 3×104 A498 and 20×104 Caki-1 cells added to the upper chamber of the transwell for analysis of migration. To assess invasion, 6×104 A498 and 40×104 Caki-1 cells were seeded in chambers where Matrigel (Thermo Fisher Scientific; Waltham, USA) had been added in advance. After 24 hours (A498 cells) or 48 hours (Caki-1 cells) incubation, the invaded cells were fixed in 100% methanol, then stained with 0.05% crystal violet before counting of 5 randomly selected fields.
Bioinformatics analysis
We analyzed USP2 mRNA expression in four renal statistics, including Jones, Eroukhim, Gumz and Lenburg renal statistics using datasets downloaded from Oncomine (https://www.oncomine.org). A total of 533 ccRCC cases, including 72 paired cases, and clinical data on age, gender, T, N, G, and TNM stages, as well as metastasis, recurrence, overall survival (OS) and disease-free survival (DFS) of patients in TCGA-KIRC database, were retrieved from the cBioPortal (http://www.cbioportal.org/public-porta). The paired normal cases were derived from normal or adjacent renal tissues of corresponding ccRCC patients and used as a control for ccRCC tissues. The gene set enrichment analysis (GSEA) platform was used to assess pathways enriched in the gene set, based on the pathway Enrichment Score (ES).
Statistical analysis
All statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software, Inc., USA) and SPSS Statistics 22.0 software (IBM SPSS, Chicago, IL). Data on paired cases were analyzed using a paired sample t-test, while analysis of unpaired cases was performed using a one-way analysis of variance (ANOVA) or t-test. In addition, a Pearson correlation coefficient was used to assess the relationship between two factors. The Kaplan-Meier analysis was performed to estimate the correlation between USP2 expression with overall survival (OS) and disease-free survival (DFS) times with the log-rank test. To evaluate the diagnostic value of USP2 mRNA expression in ccRCC patients, we generated receiver operating characteristic (ROC) curves and area under the curve (AUC). Finally, prognostic significance of USP2 in ccRCC was analyzed by univariate and multivariate Cox proportional hazard regressions. All in vitro experiments were performed in triplicates and all data represented as mean ± SEM. A confidence threshold, P<0.05, was used to analyze statistical significance. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Results
USP2 is significantly downregulated in ccRCC clinical samples and cell lines
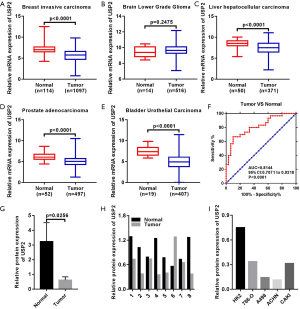
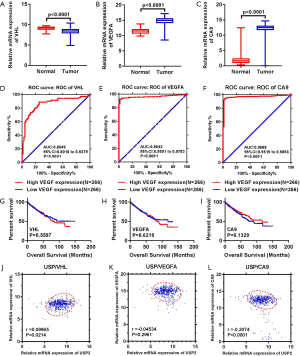
Differential expression of USP2 mRNA, in ccRCC, was first evaluated by analyzing four data sets from the Oncomine database. Results revealed reduced expression levels of USP2 in tumor tissues compared to adjacent renal tissues (Figure 1A,B,C,D). This reduction in expression was confirmed by the TCGA-KIRC database, in which 533 cases, including 72 paired cases (the paired normal cases were derived from normal kidney tissue or adjacent renal tissue of the corresponding ccRCC patients as a control for ccRCC tissue) were analyzed (Figure 1E,F). Moreover, the aberrant expression of USP2 mRNA in other tumors, including breast invasive carcinoma, brain lower grade glioma, liver hepatocellular carcinoma, prostate adenocarcinoma and bladder urothelial carcinoma, were also evaluated by analyzing datasets from the TCGA database (Figure S1A,B,C,D,E). To further verify the results from bioinformatics analysis, we performed a quantitative real-time polymerase chain reaction (qRT-PCR) analysis on 30 pairs of clinical samples collected between 2017–2018. Here, we found significantly lower levels of USP2 mRNA in ccRCC tissues relative to those in paired adjacent renal tissues (Figure 1G,H). To further confirm this aberrant expression, we constructed ROC curves based on USP2 expression in 30 pairs of clinical tissues. Results from clinical data showed that USP2 expression was able to differentiate ccRCC from normal tissues with an area under the curve (AUC) of 0.8144 (95% CI: 0.7071 to 0.9218, P<0.0001) (Figure S1F). An immunohistochemistry (IHC) assay performed to evaluate USP2 expression showed a similar profile (Figure 1I). In addition, we investigated USP2 protein expression using western blot analysis in ccRCC and adjacent renal tissues, and found low expression of the gene in tumor tissues (Figure 1J). As expected, USP2 protein expression in all the ccRCC cells (786-0, A498, ACHN, CAKI) was reduced relative to the control cell line (HK2), consistent with the previous results (Figure 1K). Furthermore, we conducted quantitative analysis of the results of immunohistochemistry and western blot (Figure S1G,H,I). In summary, USP2 expression dramatically decreased in ccRCC tissues and cells compared to adjacent tissues and immortalized renal epithelial cells.
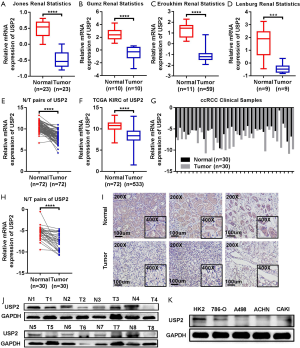
Low USP2 mRNA level is associated with various clinicopathological variables and poor prognosis in ccRCC patients
To understand the relationship between levels of USP2 expression and clinicopathological variables in ccRCC patients, we performed a more in-depth analysis of the TCGA database (Table 1). The resulting data showed a lower mRNA expression in male than in female patients (Figure 2A). Similarly, USP2 downregulation correlated with overall survival status, Grade stage, renal tumor size, weight and malignancy in ccRCC (Figure 2B,C,D,E). On the other hand, USP2 mRNA levels were negatively correlated with the T, TNM and G stages after a one-way ANOVA (P<0.05, Figure 2F,G,H). In addition, differences between cancer and adjacent renal tissues with metastasis or non-metastasis were also exhibited (Figure 2I). However, no association was found between levels of USP2 mRNA expression with age (Figure S2A).
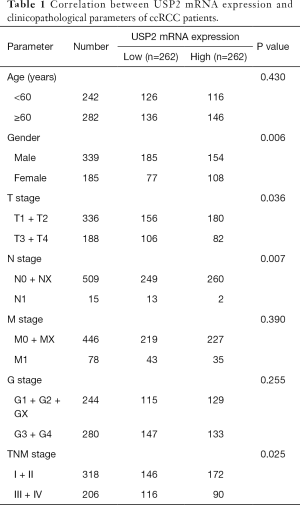
Full table
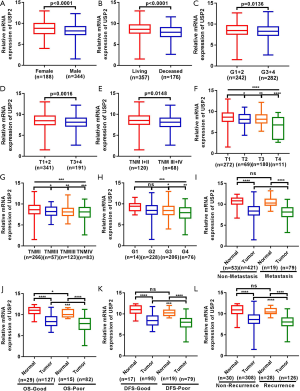
To explore the role of USP2 in differentiating benign and malignant prognosis, we performed individual analysis based on clinical outcomes from the TCGA database. Clinical outcomes of 533 cases, including 72 paired cases, were classified as OS-good (≥5 years, alive) and OS-poor (≤2 years, dead) according to the overall survival (OS) time and overall survival status, and then the USP2 mRNA levels in normal and tumor tissues of corresponding patients were analyzed. Similarly, the classification of DFS-good (≥5 years, disease-free) and DFS-poor (≤2 years, recurred/progressed) was based on disease-free survival (DFS) time and disease-free survival status. Results revealed a significant decline in USP2 expression in cancer and normal tissues of OS-poor patients compared to OS-good patients (Figure 2J). Similarly, DFS-poor patients showed much lower USP2 levels in cancer tissues compared to DFS-good ones (Figure 2K), and ccRCC patients with or without recurrence in cancer tissues could be well classified by the mRNA levels (Figure 2L). In summary, these results suggest that low USP2 levels are indicative of a significant risk of high stage. Consequently, USP2 could play a role as a potential biomarker and an independent prognosis- or clinical outcome-related classifier for ccRCC.
Downregulation of USP2 correlated with a shorter prognosis time of ccRCC patients
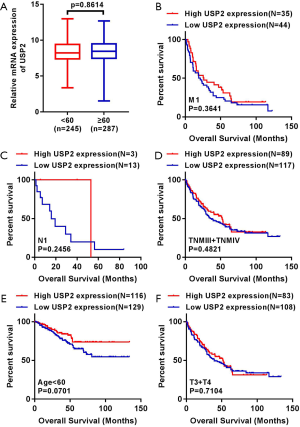
We generated Kaplan-Meier curves to investigate the correlation between mRNA levels of USP2 and prognosis time of patients. Based on the median value from USP2 expression, 532 ccRCC patients from the TCGA-KIRC database were divided into ‘high’ and ‘low’ groups. We found a shorter overall survival time in patients in the group with low USP2 levels (Figure 3A, log-rank test, P=0.0009). Subsequently, we performed an overall survival analysis to understand USP2 mRNA levels in subgroups of ccRCC patients with results providing further evidence that low expression is important in prognosis of ccRCC in male patients (Figure 3B, P=0.0062), age ≥60 years (Figure 3C, P=0.0019), T1+T2 stage (Figure 3D, P=0.0003), N0 stage (Figure 3E, P=0.0370), non-metastasis (Figure 3F, P=0.0023), TNM (I + II) (Figure 3G, P=0.0005), G1+G2 stage (Figure 3H, P=0.0285) and G3 + G4 stage (Figure 3I, P=0.0227), but not with M1 stage, N1 stage, TNM (III + IV), age <60 years, T3 + T4 stage (Figure S2B,C,D,E,F).
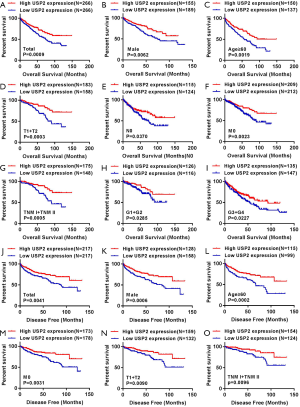
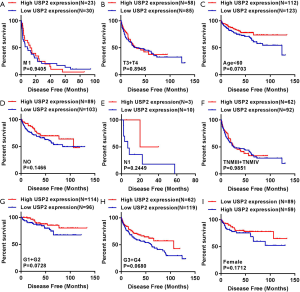
To further verify the prognostic value of USP2, we used Kaplan-Meier curves to analyze the relationship between USP2 mRNA levels and disease-free survival (DFS) time. Here, a total of 434 ccRCC patients were divided into ‘low’ and ‘high’ group using a similar classification method described above. Results indicated a significantly higher DFS in the group with high USP2 expression compared to that with low expression (Figure 3J, log-rank test, P=0.0041). In addition, we analyzed the DFS in subgroups of ccRCC patients, and found that low USP2 expression can also be a prognostic factor for patients with male (Figure 3K, P=0.0006), age ≥60 years (Figure 3L, P=0.0002), non-metastasis (Figure 3M, P=0.0031), T1 + T2 stage (Figure 3N, P=0.00901), TNM (I+II) (Figure 3O, P=0.0096) in ccRCC. However, this was not the case with M1, T3 + T4 stage, age <60 years, N0 and N1, TNM (III + IV), G1 + G2 and G3 + G4 stages, as well as in females (Figure S3).
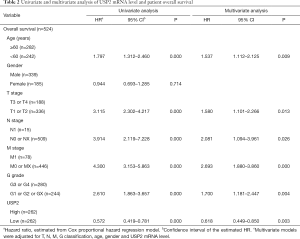
Additionally, the prognostic value of USP2 in ccRCC was further evaluated by the Cox regression model in which OS time and OS status, or DFS time and DFS status, were considered as dependent variables. The status of USP2 expression in ccRCC risk factors were assessed by univariate and multivariate analysis, which revealed that USP2 was an independent prognostic factor for ccRCC patients (Tables 2 and 3). And multivariate models were adjusted for age, gender, T stage, N stage, M stage, G grade and USP2 mRNA level.
Full table
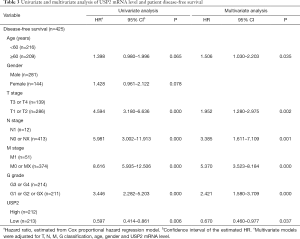
Full table
Diagnostic value of USP2 mRNA expression in ccRCC patients
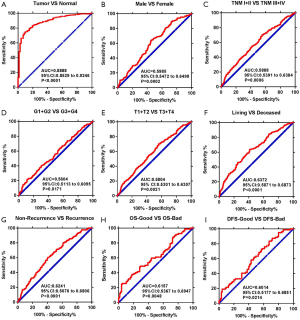
To test whether USP2 has diagnostic value in ccRCC, receiver operating characteristic (ROC) curves for the clinicopathological variables were analyzed. Results showed that ccRCC could be sensitively differentiated from normal tissues by USP2 expression, with an AUC of 0.8888 (95% CI: 0.8529 to 0.9246; P<0.0001) (Figure 4A). A further analysis of USP2 mRNA levels in subgroups of ccRCC patients, including gender, T/N/G/TNM stage, metastasis/OS/DFS status, OS-good/poor and DFS-good/poor, showed that low mRNA expression had an effective diagnostic value in ccRCC patients with male vs. female (AUC =0.5985, P=0.002), TNM (I + II) vs. (III + IV) stage (AUC =0.5888, P=0.006), (G1 + G2) vs. (G3 + G4) stage (AUC =0.5604, P=0.1771), (T1 + T2) vs. (T3 + T4) stage (AUC =0.5804, P=0.0021), OS living vs. deceased status (AUC =0.6372, P<0.0001), non-recurrence vs. recurrence (AUC =0.6241, P<0.0001), OS-good vs. OS-poor (AUC =0.6157, P=0.0048), DFS-good vs. DFS-poor (AUC =0.6014, P=0.0214) (Figure 4B,C,D,E,F,G,H,I).
In order to further investigate the clinical value of USP2 in ccRCC, we compared it with existing markers. The three main predictive and prognostic markers validated in RCC are Von Hippel Lindau (VHL), vascular endothelial growth factor (VEGF) and carbonic anhydrase IX (CAIX) (25,26). We analyzed datasets from the public database, TCGA, for differential expression of VHL, VEGFA, CAIX, and assessed their prognostic and diagnostic value by Kaplan-Meier curves and ROC curves (Figure S4A,B,C,D,E,F,G,H,I). Results showed USP2 expression had similar diagnostic value, with an AUC of 0.8888, compared with VHL (AUC =0.8649, P<0.0001), VEGFA (AUC =0.9642, P<0.0001), CAIX (AUC:0.9688, P<0.0001). However, no association was found between OS time and mRNA levels of VHL (P=0.5597), VEGFA (P=0.6210), CAIX (P=0.1329). Therefore, in comparison to these existing genes, USP2 has a comparative advantage in predicting the prognosis of ccRCC patients by its mRNA levels. Moreover, USP2 was found to be related to CAIX (P<0.0001) and VHL (P=0.0214), but not with VEGFA (P=0.2961) (Figure S4J,K,L). In conclusion, quantification of USP2 gene expression, coupled with a crucial diagnostic and prognostic value, could be used as an important diagnostic marker and an independent factor for ccRCC patients, in addition to VHL, VEGFA and CAIX.
Overexpression of USP2 significantly represses proliferation, migration, and invasion of ccRCC
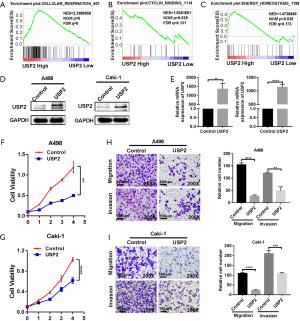
To investigate the role of USP2 in carcinogenesis and progression of ccRCC, we used Gene Set Enrichment Analysis (GSEA), based on the TCGA database, and revealed the biological pathways regulated by the gene. We found a high association between USP2 with signatures of cellular respiration signaling (NSE =2.3989558, NOM P=0, FDR q=0), cyclin-related signaling (NES =1.5381801, NOM P=0.029, FDR q=0.131) and energy homeostasis (NES =1.4738845, NOM P=0.035, FDR q=0.173), which proved to be the basis of cell growth and survival (Figure 5A,B,C). Based on these findings, we speculated that dysregulation of USP2 in ccRCC might have an important impact on ccRCC progression. To test this hypothesis, we overexpressed USP2 by transfecting plasmids into CAKi-1 and A498 cell lines (Figure 5D,E). We observed that USP2 overexpression significantly repressed the proliferation rates of A498 and CAKi-1 cells (Figure 5F,G). We then assessed migration and invasion abilities of ccRCC cells, hallmarks of tumor progression, using Transwell assays and found a significant inhibition of these processes in cells overexpressing USP2 (Figure 5H,I). Overall, proliferation, migration, and invasion of ccRCC cells in vitro were repressed by USP2 overexpression.
Discussion
Numerous efforts have been devoted to the research of genomics, transcriptomics, and proteomics in the malignant tumors (27), especially RCC. Increased understanding of the underlying molecular pathways involved in pathogenesis of RCC has contributed to development of targeted therapies (28). Currently, molecular-targeted therapies are becoming a possible option for patients with unresectable RCC (29). For instance, the targeted agents mainly refer to inhibition of VEGFR, mTORC1, c-MET and FGFR, cytokines and PD1/PDL1 immune checkpoint inhibition (27). However, most ccRCC patients still develop poor clinical progression due to low complete response rates and high drug resistance. Meanwhile, despite advanced imaging techniques have greatly improved the early detection of ccRCC patients, characterization of small renal masses and residual tumor remains a diagnostic conundrum (30). Unfortunately, reliable biomarkers and prognostic indicators for better predicting disease course are still lacking. USP2 has emerged as an oncology target in many solid malignancies, and its therapeutic potential is attracting increasing interest. However, the role of this target in renal cell carcinoma has not been elucidated. In the current work, therefore, we focused this crucial cell-growth-related gene, which is also closely correlated to cellular respiration and energy homeostasis in ccRCC cell lines. We explored its role in occurrence and progression of ccRCC, through bioinformatics analysis, and assessed its importance as a diagnostic biomarker and an independent prognosis factor for ccRCC patients. In addition, we also analyzed the GeneCards and HPA RNA-seq normal tissues from PubMed in order to evaluate the value of USP2 in liquid biopsy. Results showed that the systemic expression of USP2, including in blood and other body fluids, might represent a potential biomarker in liquid biopsy (Figure S5).

Previous studies have described USP2’s aberrant expression in different tumors as well as the modification functions it plays in regulation of signaling pathways (9,31). Particularly, USP2 is highly expressed in prostate (19), and breast cancer (32), gliomas (33), and low lowly in pancreas, colon and bladder cancer (34). For instance, compared with non-muscle-invasive bladder cancer, the expression of USP2a gene in muscle-invasive bladder cancer was found to be decreased by 36.3% (34). To date, this is the first time USP2 has been detected, shown to be lowly expressed and playing a tumor-suppressive effect in kidney cancer. Previous studies show that USP2 mainly involved in the regulation of the p53 and TNF-α/NF-κB signaling pathways. It is universally acknowledged that tumor suppressor protein, p53, which is mutated in more than 50% of solid tumors (35), plays an important role in tumor inhibition. By stabilizing the activity of MDM2 and MDMX, USP2a remarkably regulated the p53 pathway, thereby significantly influencing cell survival (20,36). In addition, NF-κB has been characterized as a crucial activator of anti-apoptotic genes during TNF-α signaling and is a vital regulator during cellular processes (37,38). Targeting the NF-κB signaling pathway, therefore, may be an effective method for treatment of carcinomas and inflammatory diseases (39). USP2 was identified to positively regulate TNF-α/NF-κB signaling, with its downregulation dramatically inhibiting NF-κB activity in breast carcinomas (40). Therefore, we hypothesized that downregulation of USP2 in clinical samples as well as cell lines, was likely to affect cell migration, invasion, and proliferation through regulation of the aforementioned signaling pathways in ccRCC cases.
The human genome encodes approximately 100 deubiquitinating enzymes (DUBs), 79 of which are predicted to be active (41). To date, DUBs have been shown to be highly important in regulating cell survival and apoptosis, and are therefore regarded as candidate target strategies for medical treatment (5). USP2, a DUB that removes Ub-chains thereby stabilizing a range of substrates such as cyclin A1 (8), cyclin D1 (9), Aurora-A (42), RIP1 and TRAF2 (22,23), has been implicated in various neoplastic and non-neoplastic diseases. In addition, ccRCC is well known for mutation of the von Hippel-Lindau (VHL) tumor suppressor gene (43) and subsequent disorder of hypoxia-inducible factor (HIF) (44). Targeting the VHL/HIF pathway in advanced ccRCC, is therefore, the new therapeutic strategy and has partially been clinically successful (45). According to the findings of this study, we hypothesized that USP2 might play a regulatory role in the VHL/HIF pathway by regulating deubiquitination in ccRCC. However, the specific mechanism of action between USP2 and the pathway in ccRCC, as well as in vivo studies and knockout experiments, were not investigated and therefore forms the basis of our further research.
Since DUBs regulate numerous key pathways in cancer cells and are potentially druggable, interests in developing DUB and their inhibitors as antitumor drugs have substantially increased (24,46). The crystal structures of USP2 have been resolved in previous studies, providing a basis for future works that aim to elucidate the molecular recognition of its activities (47,48) and the potential druggable values. Despite considerable progress in the study of ubiquitin conjugation, research on DUBs is still in its infancy. Nowadays, increasing attention on DUB activator have been gained (49). In conclusion, an understanding of the functions and mechanisms of USP2 action might contribute to the discovery of novel molecular-based target therapies and that could develop a cure for ccRCC patients.
Acknowledgments
Funding: This work was supported by Key Research and Development Plan in China [Grant No. 2017YFB1303100], the National Natural Science Foundation of China (Grant No.81874090, 81672528, 81672524, 81972630, 81773282), Hubei Provincial Natural Science Foundation of China (2018CFA038), application project of the Wuhan Science and Technology Bureau (2016060101010053), Independent Innovation Foundation of Huazhong University of Science and Technology (118530309), Clinical Research Physician Program of Tongji Medical College, Huazhong University of Science and Technology (5001530015), and the Integrated Innovation Team for Major Human Disease Program of Tongji Medical College, Huazhong University of Science and Technology.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the Institutional Review Board of Huazhong University of Science and Technology (S065).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet 2009;373:1119-32. [Crossref] [PubMed]
- Moch H, Cubilla AL, Humphrey PA, et al. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur Urol 2016;70:93-105. [Crossref] [PubMed]
- Liu Z, Zanata SM, Kim J, et al. The ubiquitin-specific protease USP2a prevents endocytosis-mediated EGFR degradation. Oncogene 2013;32:1660-9. [Crossref] [PubMed]
- Singhal S, Taylor MC, Baker RT. Deubiquitylating enzymes and disease. BMC Biochem 2008;9 Suppl 1:S3. [Crossref] [PubMed]
- Lin H, Keriel A, Morales CR, et al. Divergent N-terminal sequences target an inducible testis deubiquitinating enzyme to distinct subcellular structures. Mol Cell Biol 2000;20:6568-78. [Crossref] [PubMed]
- Zhu HQ, Gao FH. The Molecular Mechanisms of Regulation on USP2's Alternative Splicing and the Significance of Its Products. Int J Biol Sci 2017;13:1489-96. [Crossref] [PubMed]
- Davis MI, Pragani R, Fox JT, et al. Small Molecule Inhibition of the Ubiquitin-specific Protease USP2 Accelerates cyclin D1 Degradation and Leads to Cell Cycle Arrest in Colorectal Cancer and Mantle Cell Lymphoma Models. J Biol Chem 2016;291:24628-40. [Crossref] [PubMed]
- Scoma HD, Humby M, Yadav G, et al. The de-ubiquitinylating enzyme, USP2, is associated with the circadian clockwork and regulates its sensitivity to light. PLoS One 2011;6:e25382. [Crossref] [PubMed]
- Pouly D, Chenaux S, Martin V, et al. USP2-45 Is a Circadian Clock Output Effector Regulating Calcium Absorption at the Post-Translational Level. PLoS One 2016;11:e0145155. [Crossref] [PubMed]
- Yang Y, Duguay D, Fahrenkrug J, et al. USP2 regulates the intracellular localization of PER1 and circadian gene expression. J Biol Rhythms 2014;29:243-56. [Crossref] [PubMed]
- Yang Y, Duguay D, Bedard N, et al. Regulation of behavioral circadian rhythms and clock protein PER1 by the deubiquitinating enzyme USP2. Biol Open 2012;1:789-801. [Crossref] [PubMed]
- Kitamura H, Ishino T, Shimamoto Y, et al. Ubiquitin-Specific Protease 2 Modulates the Lipopolysaccharide-Elicited Expression of Proinflammatory Cytokines in Macrophage-like HL-60 Cells. Mediators Inflamm 2017;2017:6909415.
- He X, Li Y, Li C, et al. USP2a negatively regulates IL-1beta- and virus-induced NF-kappaB activation by deubiquitinating TRAF6. J Mol Cell Biol 2013;5:39-47. [Crossref] [PubMed]
- Engel E, Viargues P, Mortier M, et al. Identifying USPs regulating immune signals in Drosophila: USP2 deubiquitinates Imd and promotes its degradation by interacting with the proteasome. Cell Commun Signal 2014;12:41. [PubMed]
- Nelson JK, Sorrentino V, Avagliano TR, et al. The Deubiquitylase USP2 Regulates the LDLR Pathway by Counteracting the E3-Ubiquitin Ligase IDOL. Circ Res 2016;118:410-9. [Crossref] [PubMed]
- Pouly D, Debonneville A, Ruffieux-Daidie D, et al. Mice carrying ubiquitin-specific protease 2 (Usp2) gene inactivation maintain normal sodium balance and blood pressure. Am J Physiol Renal Physiol 2013;305:F21-30. [Crossref] [PubMed]
- Graner E, Tang D, Rossi S, et al. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell 2004;5:253-61. [Crossref] [PubMed]
- Stevenson LF, Sparks A, Allende-Vega N, et al. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J 2007;26:976-86. [Crossref] [PubMed]
- Wang CL, Wang JY, Liu ZY, et al. Ubiquitin-specific protease 2a stabilizes MDM4 and facilitates the p53-mediated intrinsic apoptotic pathway in glioblastoma. Carcinogenesis 2014;35:1500-9. [Crossref] [PubMed]
- Mahul-Mellier AL, Pazarentzos E, Datler C, et al. De-ubiquitinating protease USP2a targets RIP1 and TRAF2 to mediate cell death by TNF. Cell Death Differ 2012;19:891-9. [Crossref] [PubMed]
- Mahul-Mellier AL, Datler C, Pazarentzos E, et al. De-ubiquitinating proteases USP2a and USP2c cause apoptosis by stabilising RIP1. Biochim Biophys Acta 2012;1823:1353-65.
- Xiao Z, Zhang P, Ma L. The role of deubiquitinases in breast cancer. Cancer Metastasis Rev 2016;35:589-600. [Crossref] [PubMed]
- Stillebroer AB, Mulders PF, Boerman OC, et al. Carbonic anhydrase IX in renal cell carcinoma: implications for prognosis, diagnosis, and therapy. Eur Urol 2010;58:75-83. [Crossref] [PubMed]
- Sun M, Shariat SF, Cheng C, et al. Prognostic factors and predictive models in renal cell carcinoma: a contemporary review. Eur Urol 2011;60:644-61. [Crossref] [PubMed]
- Li QK, Pavlovich CP, Zhang H, et al. Challenges and opportunities in the proteomic characterization of clear cell renal cell carcinoma (ccRCC): A critical step towards the personalized care of renal cancers. Semin Cancer Biol 2019;55:8-15. [Crossref] [PubMed]
- Seront E, Machiels JP. Targeted therapies in the treatment of advanced renal cell carcinoma. Recent Pat Anticancer Drug Discov 2009;4:146-56. [Crossref] [PubMed]
- Kotecha RR, Motzer RJ, Voss MH. Towards individualized therapy for metastatic renal cell carcinoma. Nat Rev Clin Oncol 2019;16:621-33. [Crossref] [PubMed]
- Rossi SH, Prezzi D, Kelly-Morland C, et al. Imaging for the diagnosis and response assessment of renal tumours. World J Urol 2018;36:1927-42. [Crossref] [PubMed]
- Tomala MD, Magiera-Mularz K, Kubica K, et al. Identification of small-molecule inhibitors of USP2a. Eur J Med Chem 2018;150:261-7. [Crossref] [PubMed]
- He J, Lee HJ, Saha S, et al. Inhibition of USP2 eliminates cancer stem cells and enhances TNBC responsiveness to chemotherapy. Cell Death Dis 2019;10:285. [Crossref] [PubMed]
- Tao BB, He H, Shi XH, et al. Up-regulation of USP2a and FASN in gliomas correlates strongly with glioma grade. J Clin Neurosci 2013;20:717-20. [Crossref] [PubMed]
- Jeong P, Ha YS, Yun SJ, et al. Assess the expression of ubiquitin specific protease USP2a for bladder cancer diagnosis. BMC Urol 2015;15:80. [Crossref] [PubMed]
- Nigro JM, Baker SJ, Preisinger AC, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature 1989;342:705-8. [Crossref] [PubMed]
- Allende-Vega N, Sparks A, Lane DP, et al. MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene 2010;29:432-41. [Crossref] [PubMed]
- Van Antwerp DJ, Martin SJ, Kafri T, et al. Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science 1996;274:787-9. [Crossref] [PubMed]
- Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science 1996;274:782-4. [Crossref] [PubMed]
- Sarkar FH, Li Y, Wang Z, et al. NF-kappaB signaling pathway and its therapeutic implications in human diseases. Int Rev Immunol 2008;27:293-319. [Crossref] [PubMed]
- Metzig M, Nickles D, Falschlehner C, et al. An RNAi screen identifies USP2 as a factor required for TNF-alpha-induced NF-kappaB signaling. Int J Cancer 2011;129:607-18. [Crossref] [PubMed]
- Sowa ME, Bennett EJ, Gygi SP, et al. Defining the human deubiquitinating enzyme interaction landscape. Cell 2009;138:389-403. [Crossref] [PubMed]
- Shi Y, Solomon LR, Pereda-Lopez A, et al. Ubiquitin-specific cysteine protease 2a (USP2a) regulates the stability of Aurora-A. J Biol Chem 2011;286:38960-8. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43-9. [Crossref] [PubMed]
- Kaelin WG. Von Hippel-Lindau disease. Annu Rev Pathol 2007;2:145-73. [Crossref] [PubMed]
- Srinivasan R, Ricketts CJ, Sourbier C, et al. New strategies in renal cell carcinoma: targeting the genetic and metabolic basis of disease. Clin Cancer Res 2015;21:10-7. [Crossref] [PubMed]
- Nicholson B, Marblestone JG, Butt TR, et al. Deubiquitinating enzymes as novel anticancer targets. Future Oncol 2007;3:191-9. [Crossref] [PubMed]
- Renatus M, Parrado SG, D'Arcy A, et al. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure 2006;14:1293-302. [Crossref] [PubMed]
- Leung I, Dekel A, Shifman JM, et al. Saturation scanning of ubiquitin variants reveals a common hot spot for binding to USP2 and USP21. Proc Natl Acad Sci U S A 2016;113:8705-10. [Crossref] [PubMed]
- Song Y, Li S, Ray A, et al. Blockade of deubiquitylating enzyme Rpn11 triggers apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Oncogene 2017;36:5631-8. [Crossref] [PubMed]


