
Conjoint analysis of lncRNA and mRNA expression in rotator cuff tendinopathy
Introduction
Rotator cuff tendinopathy (RCT), previously commonly described as “tendonitis” or “tendinitis”, is a common musculoskeletal disorder and accounts for 85% of shoulder pain in patients (1). In clinic settings, RCT is easily overlooked, even by experienced physicians because of the poor relationship between structural failure and symptoms, and this worsens the treatment outcomes (2). Furthermore, despite the advances in understanding RCT and innovations in surgical techniques, conservative treatments have limited efficacy and the RC re-tear rate after surgical repair is still high due to a poor understanding of the occurrence mechanism of RCT (3). Therefore, potential factors that trigger RCT and its therapeutic targets should be identified.
Histological studies have shown the thinning and disorientation of fibers, degeneration of hyaline, increased level of ground substances, deposition of adipocytes between fibers, and relative lack of inflammatory cells in RCT (4). These findings suggest that the mechanism of RCT might involve a combination of extrinsic, environmental, and, especially, intrinsic factors (2). However, key pathways associated with RCT have not been elucidated, and there is no general consensus on the mechanisms involved. Studies on the molecular mechanisms of RCT have largely focused on a small number of genes in an attempt to provide complete information on the pathogenesis. Numerous mRNAs involved in RCT have been identified, and this has helped elucidate the underlying mechanisms (5,6).
Long non-coding RNAs (lncRNAs), which are transcripts longer than 200 bp and do not encode proteins, are regarded as key regulators of diverse biological pathways and cellular processes across all branches of life (7). Overexpression, deficiency, or mutation of lncRNAs has been implicated in numerous human diseases (8,9), such as cancer and systemic diseases (10). Although the functions of the majority of lncRNAs are unknown, some classically defined lncRNAs have unique functions in regulating life activities (11).
LncRNAs involved in tendon and muscle differentiation and regeneration have been reported. LncH19 gives rise to miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration (12). H19 accelerates tenogenic differentiation and promotes tendon healing by targeting miR-29b-3p and activating TGF-β1 signaling in humans (13). Linc-MD1, a muscle-specific lncRNA, governs the time of muscle differentiation by acting as a competing endogenous RNA (ceRNA) in mouse and human myoblasts (14). These findings suggest that other lncRNAs, which play significant roles in the pathogenesis of RCT, may exist. Therefore, in this study, we subjected samples obtained from patients with RCT and normal tendons (NTs) to lncRNA and mRNA profiling by RNA sequencing (RNA-Seq) in order to identity potential roles and mechanisms of lncRNAs and mRNAs in RCT.
Methods
Patient samples, ethics, and clinical data
All experiments and the study protocols, including sample acquisition and analysis, were approved by the institutional review board of the Human Research Ethics Committee of Army Military Medical University (approval no.: KY201838). Ten patients were carefully selected. Patients who had type 1 and 2 diabetes, hypertension, hyperlipidemia, tendon infection, smoke, alcohol addiction, previous injury and surgery of RC tendon were excluded from the study. Informed consent was obtained from all study subjects.
After removing the synovium tissue and scar tissue at the edge of the torn RC tendon, two tendon samples of approximately 2 mm × 2 mm size were collected using a pair of arthroscopic scissors. One sample was immediately frozen in liquid nitrogen for RNA-Seq and the other was fixed with formaldehyde for histological analysis. All RCT and NT samples were confirmed by histological staining after the initial imaging and observation by arthroscopy. All pathological tendons were matched with tendon samples for patient age, sex, body mass index (BMI), and Bonar score (15) (Table 1).
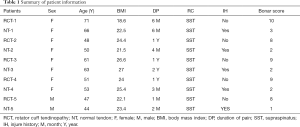
Full table
Hematoxylin and eosin (HE) staining
For histological analysis, RC tendons were placed in 4% buffered formalin. Serial sagittal paraffin sections were prepared by embedding the samples in paraffin and cutting into 4-µm thick sections. Four sections per sample were stained with HE and observed under the OLYMPUS EX-51 microscope (Tokyo, Japan) at 100× magnifications.
RNA isolation, library construction, and sequencing
The total RNA was isolated from the tendons using the RNeasy Mini Kit (Qiagen, Düsseldorf, Germany) according to the manufacturer’s instructions. RNA concentration and quality were determined using the Qubit® 2.0 Fluorometer (Life Technologies, USA) and Nanodrop One spectrophotometer (Thermo Fisher Scientific, USA). Samples with an RNA integrity number of above 7.0 were used for sequencing. Paired-end libraries were synthesized using the VAHTS Total RNA-Seq (H/M/R) Library Prep Kit (Vazyme, Nanjing, China) per the manufacturer’s instructions. Briefly, after the removal of rRNA, RNA was purified using magnetic beads. Then, mRNA was fragmented into small pieces by heating at 94 °C in the presence of divalent cations for 8 min. The cleaved RNA fragments were copied into the first-strand cDNA using reverse transcriptase and random primers. DNA polymerase I and RNase H were used for the second-strand cDNA synthesis. The cDNA fragments were subjected to end repair, single A base addition, adapter ligation, and purification and enrichment by polymerase chain reaction (PCR) to generate the final cDNA library. The Qubit® 2.0 fluorometer and Agilent 2100 Bioanalyzer (Agilent Technologies, USA) were used for quantifying and validating the purified libraries, respectively, confirming the insert size and calculating the molar concentration. Clusters were generated using cBot with the library diluted to 10 pM. The libraries were sequenced on the NovaSeq 6000 system (IL, USA). Library construction and sequencing were performed by Sinomics (Shanghai, China).
After performing quality control based on Q30, paired-end sequence files were mapped to the human reference genome hg38 (GRCh38) using Hisat2 software (v2.0.5). Gene abundance was visualized according to the fragments per kilobase of exon per million (FPKM) read. The fragment count within each gene was performed using StringTie software (v1.3.6) and normalized using TMM algorithm. LncRNAs and mRNAs with a fold change of ≥2 and P of ≤0.01 were considered differentially expressed.
Quantitative reverse transcription-PCR (qRT-PCR)
The original five pairs and another five pairs of tendons were used for verification. The samples were harvested and analyzed as described above. cDNA was generated from the total RNA using the Superscript III First-Strand Synthesis Kit (TaKaRa) according to the manufacturer’s instructions. qRT-PCR was conducted using the SYBR Green RT-PCR Kit (TaKaRa) and ABI Prism 7900 Sequence Detection System (PE Applied Biosystems, Foster City, CA, USA). The primers are listed in Table 2. mRNA and lncRNA expression levels were normalized to GAPDH expression. The relative changes in gene expression were calculated using the 2–∆∆Ct method (16). The analysis was conducted three times independently.
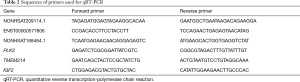
Full table
Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway analyses
The GO term enrichment analysis was used to identify potential functions of the differentially expressed mRNAs identified by RNA-Seq (17). This analysis covers three domains, including biological process, molecular function, and cellular component (http://www.geneontology.org). The KEGG pathway analysis was conducted using the KEGG database (http://www.genome.ad.jp/kegg) in R (18).
LncRNA-mRNA co-expression network analysis
Functional roles of the differentially expressed lncRNAs were predicted using the co-expression analysis. Co-expression relationships of the identified lncRNAs with mRNAs were evaluated. Pearson’s correlation between the identified lncRNAs with mRNAs was calculated, and those with a correlation coefficient of ≥0.90 and P of <0.05 were selected. The lncRNAs-mRNA interaction network was visualized using Cytoscape software (3.7.1) (19).
LncRNA-miRNA-mRNA (ceRNA) network analysis
To identify interactions between differentially expressed mRNAs and lncRNAs, lncRNA-miRNA-mRNA networks were constructed. miRNA-mRNA and miRNA-lncRNA interactions were identified using the algorithm miRanda. Network maps of the 500 lncRNA-miRNA-mRNA interactions with the highest sum of maximum energy were visualized using Cytoscape software (3.7.1).
Statistical analysis
qRT-PCR data were analyzed with the two-tailed Student’s t-test using SPSS version 22.0 (IBM Corp, Armonk, NY, USA). Differences with P<0.05 were considered statistically significant.
Results
Characteristics of RCT
RCT showed bursal involvement, fat infiltration, and collagen fiber curling in T2-weighted coronal magnetic resonance imaging when compared with RC tear caused by trauma. The RCT samples exhibited synovial hyperplasia, and they were light yellow when compared with NT tissues in arthroscopy. When dragged with a pair of arthroscopic forceps, the pathological tendon appeared stiff and less elastic.
Histologically, the collagen was arranged regularly and tightly in the NT. Tenocytes with elongated spindle-shaped nucleus were not conspicuous. However, in RCT, the collagen had lost its normal architecture and showed uneven thickness. Furthermore, there was a cluster of capillaries and the nuclei were round. Stainable mucin was present in the ground substance between discrete fiber bundles. The modified Bonar scores were calculated for each sample (Table 1, Figure 1).
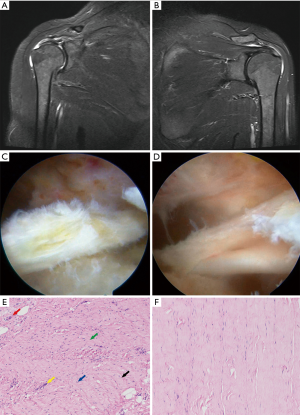
Differentially expressed lncRNAs and mRNAs in RCT
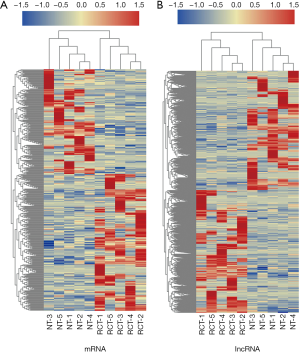
To investigate lncRNAs potentially involved in RCT, RNA-Seq was performed to identify mRNAs and lncRNAs differentially expressed among five pairs of RCT tissue and NT samples. We identified 66,596 lncRNAs and 128,852 mRNAs; 419 differentially expressed lncRNAs and 1,541 mRNAs were identified using the following criteria: fold change of >2 and P of <0.01. There were 182 upregulated and 237 downregulated lncRNAs (http://fp.amegroups.cn/cms/ee5cc717fa57c722e15644da5372c034/atm.2020.02.149-1.pdf), and 761 upregulated and 780 downregulated mRNAs among the differentially expressed RNAs (http://fp.amegroups.cn/cms/dd787cc043f1868090aac3c09c58e960/atm.2020.02.149-2.pdf). Heatmaps of the differentially expressed mRNAs and lncRNAs are presented in Figure 2.
Validation of RNA-Seq data by qRT-PCR
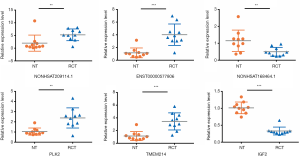
To evaluate the accuracy of the outcomes of RNA-Seq and potential targets of RCT, three significantly differentially expressed lncRNAs and three mRNAs [NONHSAT209114.1, ENST00000577806, NONHSAT168464.1, polo-like kinase 2 (PLK2), transmembrane protein 214 (TMEM214), and insulin growth factor 2 (IGF2)] were selected to verify RNA-Seq data by qRT-PCR. Ten pairs of RCT and NT samples were used for validation. Relative expression levels of all the tested RNAs were significantly different between the two groups. Furthermore, they were consistent with the RNA-Seq results, indicating that the RNA-Seq data were reliable and their potential role in RCT occurrence (Figure 3).
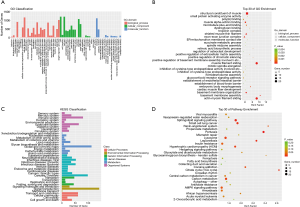
GO enrichment and KEGG pathway analysis of differentially expressed mRNAs and lncRNAs
The GO analysis was used to gain deeper insights into the biological processes that are potentially mediated by the differentially expressed mRNAs and lncRNAs in RCT. Most of the differentially expressed RNAs were involved in biological processes, whereas the less differentially expressed RNAs were assigned the GO term molecular function. Within the biological process, cellular component, and molecular function classes, most genes were associated with cellular process, cell part, and binding, respectively (Figure 4A). The mRNAs were enriched in FATZ binding, skeletal muscle thin filament assembly, muscle filament sliding, actin-myosin filament sliding, and structural muscle constituent (Figure 4B). The KEGG pathways comprised cellular processes, environmental information processing, genetic information processing, human diseases, metabolism, and organismal systems. Most genes were enriched in the immune system; global/overview metabolism; cancer; folding, sorting and degradation; signal transduction; and cellular community in the organismal systems, metabolism, human diseases, genetic information processing, environmental information processing; and cellular processes (Figure 4C). Furthermore, the KEGG pathway enrichment analysis showed that genes were enriched in the citrate cycle, renin-angiotensin system, sphingolipid signaling pathway, glyoxylate and dicarboxylate metabolism, B cell receptor signaling pathway, and p53 signaling pathway (Figure 4D).
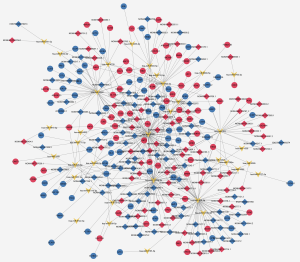
ceRNA network analysis
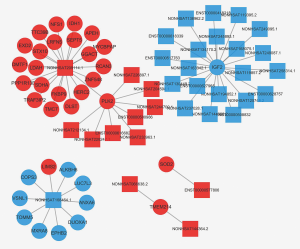
To investigate potential interactions among lncRNAs, mRNAs, and miRNAs, a ceRNA network was constructed. The network comprised 300 nodes and 442 edges, involving 139 lncRNAs, 126 mRNAs, and 35 miRNAs. The most-linked miRNA, hsa-miR-1273g-3p, had 62 edges. The most-linked lncRNA and mRNA were NONHSAT224676.1 and ARHGAP6, respectively, both with six edges (Figure 5).
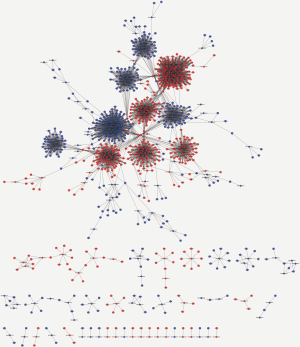
Co-expression of lncRNAs and mRNAs
To identify potential functions of the identified lncRNAs and mRNAs, the co-expression network analysis was performed. The network comprised 60 nodes and 7,702 edges, comprising 248 lncRNAs and 738 mRNAs. The most-linked lncRNA, ENST00000540997, had 99 edges. The most-linked mRNA, SPP1 had 155 edges (Figure S1). The co-expression network of six validated lncRNAs and mRNAs (NONHSAT209114.1, ENST00000577806, NONHSAT168464.1, PLK2, TMEM214, and IGF2) with DEG and DEL is shown in Figure 6. PLK2 was highly correlated with NONHSAT209114.1, according to the lncRNAs-mRNA co-expression network analysis (r=0.907, P<0.001).
Discussion
In this study, we aimed to identify key lncRNAs and mRNAs involved in RCT occurrence. By RNA-Seq, we identified 419 lncRNAs and 1,541 mRNAs differentially expressed in RCT. The differential expression of NONHSAT209114.1, ENST00000577806, NONHSAT168464.1, PLK2, TMEM214, and IGF2 was confirmed by qRT-PCR, indicating the reliability of the RNA-Seq data and the potential roles of these RNAs in RCT occurrence.
The potential action mechanism of these validated mRNAs has been reported. Upregulation of mircoRNA-27a inhibited synovial angiogenesis, damaged cartilage collagen, and alleviated inflammation via inhibition of PLK2 in rats with knee osteoarthritis (20). IGF2 and IGF1 are involved in tendon remodeling and myoblast differentiation and proliferation (21). TMEM214 is a vital mediator of endoplasmic reticulum stress-induced apoptosis (22). These differentially expressed RNAs may share familiar function in RCT occurrence.
The GO and KEGG pathway enrichment analyses indicated that several pathways, such as the citrate cycle, p53 signaling, and the renin-angiotensin system, are potentially associated with RCT. These links are supported by several previous studies. Kim et al. observed that fatty acid and amino acid oxidation metabolism via the citric acid cycle are increased in the skeletal muscle of mice after radiation (23). The number of cells expressing p53 and apoptotic tendon cells was significantly increased in partially torn supraspinatus (SST) tendons (24). Furthermore, Nyyssönen et al. reported that the use of renin-angiotensin II receptor antagonists was significantly associated with Achilles tendon rupture (25). Together, differentially expressed mRNAs associated with these pathways can help elucidate potential mechanisms of lncRNAs in RCT.
Numerous factors, including smoking (26), alcohol assumption (27), and comorbidities and education level (28), have been implicated in RCT. In addition, combinations of extrinsic and intrinsic factors have been recognized as critical causes of RCT. The extrinsic theory suggests that RCT is due to over-use, over-load, and repeated irritation. Although the subacromial impingement syndrome is widely accepted as a characteristic of RCT, it is still debated whether acromial irritation leads to RCT. Neer et al. reported that 95% of all RC pathology cases were caused by irritation from the anteroinferior aspect of the acromion to the superior aspect of the RC (29,30). However, based on ultrasound and MRI images, the association between symptoms related to RCT and structural failure is poor (31). A multicenter randomized surgical trial showed that arthroscopic subacromial decompression had no additional benefit over arthroscopy alone, but it offered better symptom improvement than no treatment, and the difference was not clinically significant (32). These findings indicate that chronic impingement may not be strongly associated with RCT occurrence. Furthermore, if impingement causes RCT, the degenerated tissue would be located in the bursal side, whereas the majority of partial-thickness tears will be observed in the joint side (33). This indicates that RCT occurrence involves intrinsic rather than extrinsic factors; however, this remains to be explored.
The intrinsic factors include original tendon pathology. Besides primary RC degeneration, RC vascularity and changes associated with ageing have garnered the attention of researchers. Lindblom supported Codman’s view of “critical portion” by showing that areas of avascularity were located near the insertion of SST (34). In addition, type III collagen is increased in RCT. As the ratio of type I to type III collagen affects RC tendon strength, RCT increases the risk of rupture (35). Exosomes from tendon stem cells can balance the synthesis and degradation of tendon extracellular matrix, thus promoting tendon healing (36). Factors contributing to this process remain to be elucidated. We found that differentially expressed genes (DEGs) in RCT were involved in VEGF signaling (enrichment factor =1.34). Genes such as COL11A1, COL2A1, and COL4A4 were differentially expressed in RCT (fold change =4.23, 2.95, and –4.40, respectively). These genes provide insights into the potential mechanism underlying the above theory.
The role of inflammation in tendinopathy was unclear. It has been shown that inflammatory cell infiltration in RCT is not to the extent observed in the classical inflammatory response (37). However, Dakin et al. concluded that chronic inflammation is a feature of Achilles tendinopathy and rupture. Achilles tendinopathy tissues were enriched in CD14+ and CD68+ cells, which were highly expressed in Achilles tendinopathy tissues. Inflammation-related pathways such as NF-κB, interferon, and STAT-6 activation, were involved in RCT, presenting a complex inflammation signature (38). In our study, GO enrichment showed that genes associated with inflammation, including the adaptive immune response and inflammatory response, were highly expressed in RCT. The KEGG pathway analysis showed that DEGs were highly enriched in the B cell receptor signaling pathway. These findings indicate that inflammation is also a feature of RCT and crucially contributes to the occurrence of RCT. Considering that lncRNAs have critical roles in various aspects of immunity, such as the production of inflammatory mediators, regulation of the innate immune response, and migration of cells by regulating protein-protein interactions (39), the roles of immune-related lncRNAs in RCT remain to be elucidated.
In this study, we first systematically evaluated lncRNA and mRNA expression profiles in patients with RCT and characterized the potential genetic pathways underlying this disorder. A systematic comparison unraveled common pathological changes in patients with RCT. However, this study had some limitations. First, RC tendon cannot be harvested from healthy individuals because of ethical principles; thus, we used samples from patients undergoing fracture surgery or suffering from other trauma events. Second, most adults may have RC degeneration to some extent. Therefore, we evaluated the samples by HE staining and imaging, and found that a part of the samples had different degrees of degeneration. Although this reflects normal physiological processes, it may influence the study outcomes. Third, as the pathological and control samples were obtained from different patients, we could not avoid interference even after grouping the patients by age, sex, BMI, and HE staining characteristics.
Conclusions
In conclusion, we identified several lncRNAs and mRNAs differentially expressed in RCT, potentially associated with the progression of the disorder. The outcomes of this study provide valuable insights into the mechanism of RCT and suggest a series of potential targets for the diagnosis and treatment, paving the way to better treatment and prevention of this disorder.
Acknowledgments
Funding: This work was supported by the Chongqing Special Support Program for postdoctoral research fellows (grant number Xm2017179); and National Natural Science Foundation of China (grant number 81572152).
Footnote
Conflict of Interests: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of Army Military Medical University (Approval No.: KY201838) and was registered in Chinese Clinical Trial Registry (File No. ChiCTR1900022467). Written informed consent was provided by the participants or their proxy in accordance with the Helsinki Declaration as revised in 2013.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ostör AJ, Richards CA, Prevost AT, et al. Diagnosis and relation to general health of shoulder disorders presenting to primary care. Rheumatology (Oxford) 2005;44:800-5. [Crossref] [PubMed]
- Lewis JS. Rotator cuff tendinopathy. Br J Sports Med 2009;43:236-41. [Crossref] [PubMed]
- Ahn JO, Chung JY, Kim DH, et al. Differences of RNA expression in the tendon according to anatomic outcomes in rotator cuff repair. Am J Sports Med 2017;45:2995-3003. [Crossref] [PubMed]
- Cipollaro L, Sahemey R, Oliva F, et al. Immunohistochemical features of rotator cuff tendinopathy. Br Med Bull 2019;130:105-23. [Crossref] [PubMed]
- Jelinsky SA, Rodeo SA, Li J, et al. Regulation of gene expression in human tendinopathy. BMC Musculoskelet Disord 2011;12:86. [Crossref] [PubMed]
- Chaudhury S, Xia Z, Thakkar D, et al. Gene expression profiles of changes underlying different-sized human rotator cuff tendon tears. J Shoulder Elbow Surg 2016;25:1561-70. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Yan Y, Xu Z, Chen X, et al. Novel Function of lncRNA ADAMTS9-AS2 in promoting temozolomide resistance in glioblastoma via upregulating the FUS/MDM2 ubiquitination axis. Front Cell Dev Biol 2019;7:217. [Crossref] [PubMed]
- Fang H, Disteche CM, Berletch JB. X inactivation and escape: epigenetic and structural features. Front Cell Dev Biol 2019;7:219. [Crossref] [PubMed]
- Maass PG, Luft FC, Bähring S. Long non-coding RNA in health and disease. J Mol Med (Berl) 2014;92:337-46. [Crossref] [PubMed]
- Wang QS, Zhou J, Li X. LncRNA UCA1 protects cardiomyocytes against hypoxia/reoxygenation induced apoptosis through inhibiting miR-143/MDM2/p53 axis. Genomics 2020;112:574-80. [Crossref] [PubMed]
- Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev 2014;28:491-501. [Crossref] [PubMed]
- Lu YF, Liu Y, Fu WM, et al. Long noncoding RNA H19 accelerates tenogenic differentiation and promotes tendon healing through targeting miR-29b-3p and activating TGF-β1 signaling. FASEB J 2017;31:954-64. [Crossref] [PubMed]
- Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011;147:358-69. [Crossref] [PubMed]
- Maffulli N, Longo UG, Franceschi F, et al. Movin and Bonar scores assess the same characteristics of tendon histology. Clin Orthop Relat Res 2008;466:1605-11. [Crossref] [PubMed]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008;3:1101-8. [Crossref] [PubMed]
- Yan Y, Xu Z, Qian L, et al. Identification of CAV1 and DCN as potential predictive biomarkers for lung adenocarcinoma. Am J Physiol Lung Cell Mol Physiol 2019;316:L630-43. [Crossref] [PubMed]
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-61. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Liu W, Zha Z, Wang H. Upregulation of microRNA-27a inhibits synovial angiogenesis and chondrocyte apoptosis in knee osteoarthritis rats through the inhibition of PLK2. J Cell Physiol 2019;234:22972-84. [Crossref] [PubMed]
- Duan C, Ren H, Gao S. Insulin-like growth factors (IGFs), IGF receptors, and IGF-binding proteins: roles in skeletal muscle growth and differentiation. Gen Comp Endocrinol 2010;167:344-51. [Crossref] [PubMed]
- Li C, Wei J, Li Y, et al. Transmembrane Protein 214 (TMEM214) mediates endoplasmic reticulum stress-induced caspase 4 enzyme activation and apoptosis. J Biol Chem 2013;288:17908-17. [Crossref] [PubMed]
- Kim EJ, Lee M, Kim DY, et al. Mechanisms of energy metabolism in skeletal muscle mitochondria following radiation exposure. Cells 2019. [Crossref] [PubMed]
- Lundgreen K, Lian Ø, Scott A, et al. Increased levels of apoptosis and p53 in partial-thickness supraspinatus tendon tears. Knee Surg Sports Traumatol Arthrosc 2013;21:1636-41. [Crossref] [PubMed]
- Nyyssönen T, Lantto I, Lüthje P, et al. Drug treatments associated with Achilles tendon rupture. A case-control study involving 1118 Achilles tendon ruptures. Scand J Med Sci Sports 2018;28:2625-9. [Crossref] [PubMed]
- Baumgarten KM, Gerlach D, Galatz LM, et al. Cigarette smoking increases the risk for rotator cuff tears. Clin Orthop Relat Res 2010;468:1534-41. [Crossref] [PubMed]
- Passaretti D, Candela V, Venditto T, et al. Association between alcohol consumption and rotator cuff tear. Acta Orthop 2016;87:165-8. [Crossref] [PubMed]
- MOON Shoulder Group, Unruh KP, Kuhn JE, et al. The duration of symptoms does not correlate with rotator cuff tear severity or other patient-related features: a cross-sectional study of patients with atraumatic, full-thickness rotator cuff tears. J Shoulder Elbow Surg 2014;23:1052-8. [Crossref] [PubMed]
- Neer CS 2nd. Anterior acromioplasty for the chronic impingement syndrome in the shoulder: a preliminary report. J Bone Joint Surg Am 1972;54:41-50. [Crossref] [PubMed]
- Neer CS 2nd. Impingement lesions. Clin Orthop Relat Res 1983.70-7. [PubMed]
- Lewis JS. Rotator cuff tendinopathy/subacromial impingement syndrome: is it time for a new method of assessment? Br J Sports Med 2009;43:259-64. [Crossref] [PubMed]
- Beard DJ, Rees JL, Cook JA, et al. Arthroscopic subacromial decompression for subacromial shoulder pain (CSAW): a multicentre, pragmatic, parallel group, placebo-controlled, three-group, randomised surgical trial. Lancet 2018;391:329-38. [Crossref] [PubMed]
- Ogata S, Uhthoff HK. Acromial enthesopathy and rotator cuff tear. A radiologic and histologic postmortem investigation of the coracoacromial arch. Clin Orthop Relat Res 1990.39-48. [PubMed]
- Lindblom K. On pathogenesis of ruptures of the tendon aponeurosis of the shoulder joint. Acta Radiol 1939;20:563-77. [Crossref]
- Riley GP, Harrall RL, Constant CR, et al. Tendon degeneration and chronic shoulder pain: changes in the collagen composition of the human rotator cuff tendons in rotator cuff tendinitis. Ann Rheum Dis 1994;53:359-66. [Crossref] [PubMed]
- Wang Y, He G, Guo Y, et al. Exosomes from tendon stem cells promote injury tendon healing through balancing synthesis and degradation of the tendon extracellular matrix. J Cell Mol Med 2019;23:5475-85. [Crossref] [PubMed]
- Fukuda H, Hamada K, Yamanaka K. Pathology and pathogenesis of bursal-side rotator cuff tears viewed from en bloc histologic sections. Clin Orthop Relat Res 1990.75-80. [PubMed]
- Dakin SG, Newton J, Martinez FO, et al. Chronic inflammation is a feature of Achilles tendinopathy and rupture. Br J Sports Med 2018;52:359-67. [PubMed]
- Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol 2014;35:408-19. [Crossref] [PubMed]


