
Prognostic significance of mutant-allele tumor heterogeneity in uterine corpus endometrial carcinoma
Introduction
Intra-tumor heterogeneity is a hallmark of cancer, which is characterized by the presence of different subpopulations of cancer cells with distinct genetic, phenotypic, or behavioral characteristics within the same tumor, as well as between a given primary tumor and its metastases (1,2). Currently, the extent of intra-tumor heterogeneity may serve as a clinically useful biomarker in the development of personalized therapies and clinical outcomes (3-5). Genetic heterogeneity is a pivotal type of intra-tumor heterogeneity and has been considered as the product of genomic instability, which results in the presence of different subclonal populations within the tumor, and likely influences the patient’s clinical course and response to therapy (6-9). Most previous investigations have been small-scale studies or consisted of only single-cell analyses. In addition, identified markers of tumor subpopulations may not generalize well across tumor types and could be difficult to standardize and quantify. Moreover, guidelines for scoring and reporting have not yet been established. Furthermore, the analyses of genetic heterogeneity have been limited with respect to sequencing depth and the number of detected somatic mutations (10). While this concept is now well established, the quantification of genetic heterogeneity to assess intra-tumor heterogeneity is still a great challenge in clinical trials and clinical practice.
Mutant-allele tumor heterogeneity (MATH) is a quantitative assessment of genetic intra-tumor heterogeneity, which is based on whole-exome sequencing (WES) of tumors and of their matched normal DNA (11). The MATH score, which is the normalized width of this distribution, is derived directly from the mixed population of mutant-allele frequencies within a tumor (11). The MATH score has been reported to be a reliable biomarker of outcomes in patients with head and neck squamous cell carcinoma (HNSCC) (11-13), breast cancer (14,15), colorectal cancer (16), and lung adenocarcinoma (17). Although studies have successfully demonstrated that intra-tumor heterogeneity assessed by MATH is a prognostic biomarker in some cancers, the prognostic significance of the MATH score in various types of tumors has not been thoroughly explored at the genomic level.
Uterine corpus endometrial carcinoma (UCEC), which has a mortality rate of approximately 20%, is the most common gynecological malignancy in the western world (18). UCEC has traditionally been classified into two types (types I and II), based on epidemiological, clinical, and endocrine characteristics (19). A number of studies have already demonstrated that UCEC is comprised of a biologically, clinically, morphologically, and genetically heterogeneous group of tumors (20-22). In 2013, The Cancer Genome Atlas (TCGA) Research Network performed a comprehensive genomic and transcriptomic analysis of UCEC, which was based on array and sequencing technologies using common histological types (23). Despite the fact that MATH is a quantitative biomarker to assess intra-tumor heterogeneity that has been applied to some tumors, the prognostic value of MATH in UCEC patients remains to be determined.
In this study, we investigated the associations of MATH with clinical, pathological, and overall survival data in UCEC patients from the TCGA database, and explored the potential clinical implications of intra-tumor heterogeneity in UCEC. Our findings suggest that the MATH score is a quantitative measure of intra-tumor heterogeneity, and could serve as a novel biomarker for improved prognosis and personalized therapy for UCEC patients.
Methods
Eligible patients and clinical variables
The publicly available clinical data used in this study were released by TCGA and the Broad Institute of MIT and Harvard. Female patients who were diagnosed with clinical stage I–IV UCEC with acquirable level-III WES data were eligible. For each patient, the following data were acquired based on the clinical file downloaded from http://gdac.broadinstitute.org/: age at initial pathological diagnosis, vital status, days to death, days to last follow-up, clinical stage, histological grade, cancer type, hormonal therapy, pregnancies, radiation therapy, targeted molecular therapy, surgical approach, and presence/absence of diabetes. Additionally, race and body mass index (BMI) were determined based on the clinical file downloaded from https://portal.gdc.cancer.gov/. The molecular type of each sample was downloaded from attachment files of the article “Integrated Genomic Characterization of Endometrial Carcinoma” (23). Since the datasets analyzed during the current study were available in TCGA databases, ethical approval or informed consent was not required.
Genomic variables and generation of MATH
Tumor-specific mutation data from WES were downloaded from the Broad Institute of MIT and Harvard (24), where WES data had been obtained. Mutation data were available for 248 of the 560 UCEC patients. The tumor MATH score for each patient was calculated following the method described by Mroz EA and Rocco JW (11). Tumor-specific mutations had already been identified at the Broad Institute of MIT and Harvard for TCGA. The steps to determine the MATH score are summarized as follows: (I) calculating the mutant-allele fraction (MAF) for each locus as the ratio of mutant reads to total reads; (II) obtaining the absolute difference of each MAF from the median MAF value, and then multiplying the median of these absolute differences by a factor of 1.4826, thus generating the median absolute deviation (MAD); and (III) calculating the MATH score as the percentage ratio of the MAD to the median of the MAFs among the tumor’s mutated genomic loci as follows: MATH = MAD/median ×100%. The patients were separated into high- and low-MATH groups by the median of the MATH scores. Somatic mutations for each patient were downloaded from the Broad Institute for the aforementioned TCGA UCEC subjects and were compared between the low-MATH group and the high-MATH group. The fraction of somatic copy number alteration (SCNA) in TCGA UCEC subjects was obtained from the cBioPortal for Cancer Genomics (http://cbioportal.org).
Statistical and bioinformatic analysis
The association between the MATH score and clinical characteristics was evaluated by one-way analysis of variance (ANOVA), with pairwise comparisons that were adjusted using the Bonferroni test. The receiver operating characteristic (ROC) curve showed the tradeoff between sensitivity and specificity at different choices of the MATH classification cutoff for the whole cohort. To adapt the data to the following analyses, we used the median MATH score of 21.425 as the cutoff point to divide the patients into two groups of equal size; this cutoff was also adopted in subgroup analyses. The correlation coefficient between MATH scores and SCNA fraction was assessed by the Spearman method. Kaplan-Meier survival curves were used to analyze the overall survival of UCEC patients, and log-rank (Mantel-Cox) tests were applied to compare the differences between subgroups. Univariate and multivariate Cox regression was employed to evaluate the prognostic value of the MATH score, where the MATH score was used both as a categorical variable and as a continuous variable, respectively. In the whole cohort, all significant parameters identified by univariate analysis have been included in multivariate analysis. Stratified multivariate analyses were performed using the variables showing P values <0.20 in the univariate analysis, in addition to other variables considered as known prognostic factors. The predictive accuracy of different Cox models was described by the area under the ROC curve (AUC) and the index of concordance (C-index). All analyses were performed using the International Business Machines Corporation statistical software program Statistical Product and Service Solutions (SPSS), version 22.0.0 (SPSS Incorporated, Chicago, IL, USA). MATH scores were calculated by R version 3.3.3 (R Foundation for Statistical Computing, Vienna, Austria). A two-sided P value <0.05 was considered to indicate statistical significance, unless otherwise indicated.
Results
MATH and baseline characteristics in UCEC
Somatic-mutation data were available for 248 of the 560 UCEC patients, and ultimately 242 MATH scores were acquired. For all 242 eligible patients, initial pathological diagnoses were made between 1995 and 2013 (median, 2009); the median follow-up was 36.05 months (range, 0.57–188.37 months), and 33 patients (13.6%) had died. Their ages ranged from 34 to 90 years and their races were defined as the following: White; Black or African American; Asian; Native Hawaiian or other Pacific Islander; and American Indian or Alaskan Native. Tumor samples and corresponding germline DNA were composed of 194 endometrioid (type-I UCEC), 43 serous (type-II UCEC), and five mixed histology cases. Using the MATH scoring method, the scores of MAFs among loci tend to be higher for heterogeneous tumors than for homogeneous tumors. As shown in Figure 1A, tumor MATH scores ranged from 0 to 75.023, with a mean ± standard deviation (SD) of 27.114±14.724 (interquartile range, 16.820–36.203). According to the median (21.425) MATH score, patients were divided into two groups: the low-MATH group and the high-MATH group, with mean ± SD of 16.485±3.456 and 37.742±13.987, respectively. The distribution of MATH scores showed a wide spectrum of allele frequencies and strong clustering of the variants at a high MATH score (Figure 1A). The box plots of MATH distribution according to different clinicopathological characteristics of tumors are displayed in Figure 1B. The distribution of MATH scores yielded considerable variation in MATH scores and indicated that UCEC has high heterogeneity.
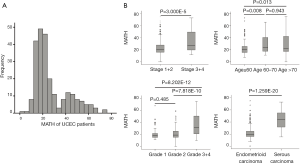
Relationships of clinical characteristics with MATH scores
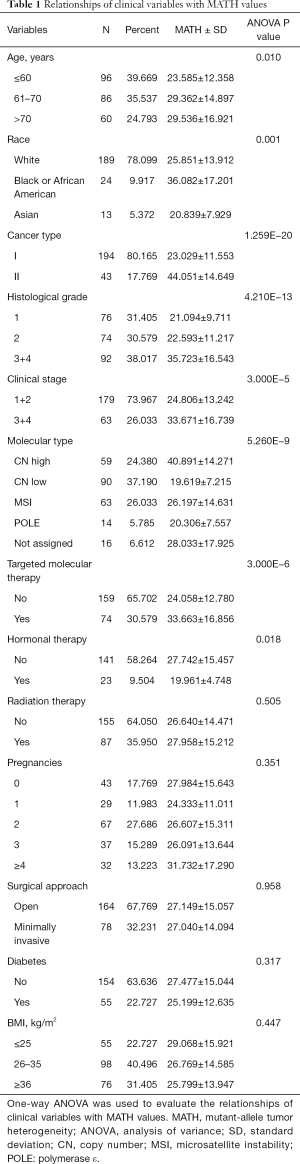
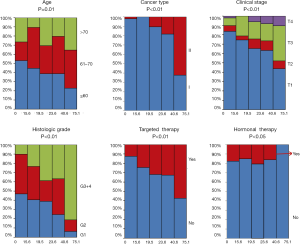
The clinical characteristics, including age, histological grade, clinical stage, and BMI, were downloaded. They are listed in Table 1. As shown in Table 1 and Figure 2, the tumor MATH scores were significantly related to age, race, cancer type, clinical stage, histological grade, molecular type, targeted molecular therapy, and hormonal therapy. In contrast, MATH scores were not significantly correlated with radiation therapy, pregnancies, surgical approach, diabetes, or BMI (Table 1). UCEC patients younger than 60 years old had low MATH scores; MATH scores of those over 60 years old were significantly higher (Table 1, Figure 2). Further analysis revealed that MATH scores were higher in type-II UCEC patients compared to those in type-I UCEC patients (P<0.01; Table 1, Figure 2). Additionally, with respect to the clinical stage, UCEC patients with tumor classification T1 and T2 had much lower MATH scores than patients with T3 and T4 (P<0.01; Table 1, Figure 2). In terms of histological grade, UCEC patients with tumor histological grade G3 and G4 tended to have significantly higher MATH scores than patients of the G1 and G2 subtypes (P<0.01; Table 1, Figure 2). Moreover, as shown in Table 1 and Figure 2, targeted molecular therapy was associated with higher MATH scores in UCEC patients (P<0.05), whereas lower MATH scores were significantly associated with menopausal hormonal therapy (P<0.05). Based on the four molecular subtypes of endometrial carcinoma, our analysis revealed that MATH scores were significantly higher in the copy number (CN) high subgroup than in other molecular subtypes (P<0.01; Table 1).
Full table
Correlation of MATH with somatic mutations and SCNA
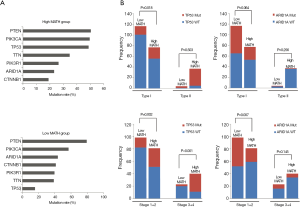
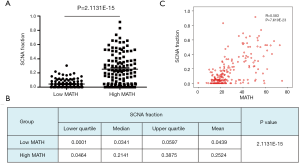
To explore the differences in somatic-mutation frequencies between the high- and low-MATH groups, the most prevalent somatic mutations in each MATH group were analyzed. As shown in Figure 3A, mutations in phosphatase and tensin homolog (PTEN), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), and phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1) were among the top five most prevalent somatic mutations in both the groups. Mutations in titin (TTN) and tumor protein p53 (TP53) were present in the top-five list only in the high-MATH group, and mutations in AT-rich interaction domain 1A (ARID1A) and catenin-beta 1 (CTNNB1) were present only in the top-five list in the low-MATH group. Therefore, TP53 and ARID1A were mutated at significantly different frequencies in the low- and high-MATH groups, as confirmed by the Pearson chi-square test and logistic regression analysis (P<0.05 in both tests). TP53 mutation rates in the low-MATH group and the high-MATH group were 15.7% and 48.8%, respectively. After adjusting variables in the logistic regression model, the difference in TP53 mutation rate was still significant between the high- and low-MATH groups (P=0.020). In the Pearson correlation analysis, TP53 mutation displayed a significantly positive correlation with type-II UCEC (P<0.001) and clinical stage (P<0.001) in UCEC patients. Further subgroup analysis was performed next. As shown in Figure 3B, a significant difference was observed in type-I UCEC patients (P=0.018) but not in type-II UCEC patients (P=0.503). With respect to the clinical stage, a similar result was found in the mutation rates of TP53 between different MATH levels within clinical stages 1–2 and clinical stages 3–4 (P=0.002 and P<0.001, respectively; Figure 3B). Another gene showing a different mutation rate in both Fisher’s exact test and logistic regression was ARID1A (P=0.001 and P=0.031), which had a significantly higher mutation rate in the low-MATH group than in the high-MATH group (43.8% and 23.1%, respectively). Additionally, we found a negative correlation between ARID1A mutation and type-I UCEC (P<0.001) or clinical stage (P<0.001). Moreover, the difference in the mutation rate of ARID1A was only significant in the clinical stage 1–2 subgroup (P=0.007), but not in the type-I subgroup (P=0.064), type-II group (P=0.256), or clinical stage 3–4 subgroup (P=0.145; Figure 3B). Next, we explored the relationship between MATH scores and SCNA. As shown in Figure 4A, UCEC patients with high MATH scores had higher SCNA fractions than those with low MATH scores. The median fractions of SCNA in the high- and low-MATH groups were 21.41% and 3.41%, respectively, and the SCNA fraction was significantly different between the high- and low-MATH groups (Figure 4B). Furthermore, Spearman’s correlation analysis revealed a significantly positive association between the MATH score and SCNA fraction, validating that the fraction of SCNA was significantly higher in the high-MATH group (Figure 4C).
Prognostic value of the MATH score for survival in UCEC patients
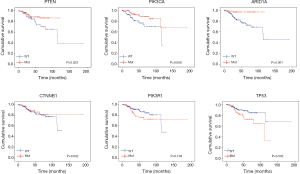
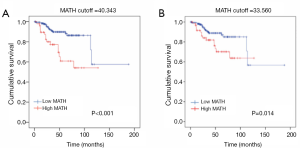
With regard to the potential prognostic value of the commonly mutated genes in UCEC, the Kaplan-Meier curves showed that patients with genetic mutations in PTEN, PIK3CA, and ARID1A exhibited longer overall survival rates than those with the wild-type genes (P<0.05; Figure 5). However, no significant difference in prognosis was observed between patients with genetic mutations in CTNNB1 and PIK3R1 and patients with their wild-type counterparts (Figure 5). In contrast, patients with TP53 mutations displayed a poorer prognosis relative to those with the wild-type TP53 gene (P<0.05; Figure 5). Survival analysis demonstrated that patients in the high-MATH group had a significantly shorter overall survival time than those in the low-MATH group based either on the median MATH score or on other MATH cutoff values (Figure 6A, Figure S1). To investigate whether the MATH score predicts somatic mutations, the relationships between the clinical outcomes of UCEC patients and the prevalent somatic mutations in the high-MATH or low-MATH groups were further analyzed. As shown in Figure 6B, ARID1A or TP53 mutations did not significantly affect prognosis in patients with high or low MATH scores. Interestingly, among patients with wild-type PTEN, patients in the low-MATH group exhibited longer overall survival rates than those in the high-MATH group (P=0.043), whereas among patients with PIK3CA mutation, low MATH scores were strongly correlated with better disease outcomes than high MATH scores (P=0.030; Figure 6B).
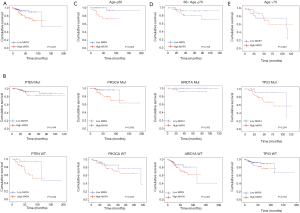
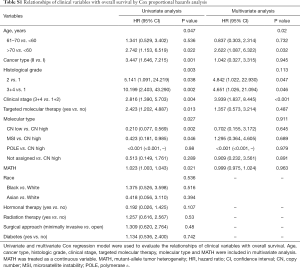
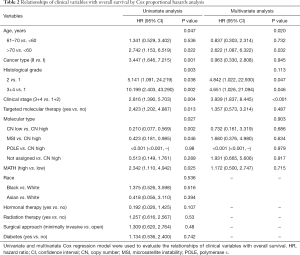
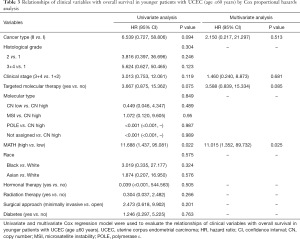
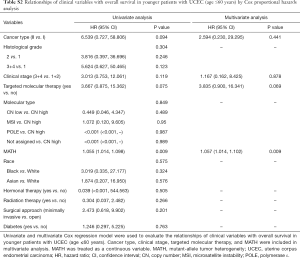
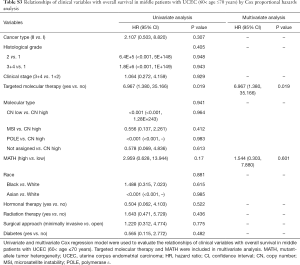
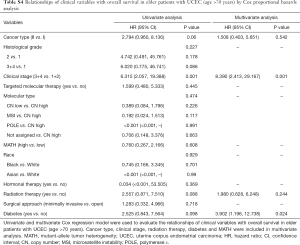
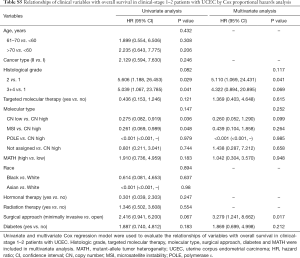
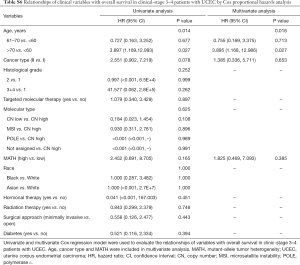
Next, we explored the prognostic value of the MATH score and other variables in all patients and subgroups. The MATH score, taken as a continuous variable, was strongly related to overall survival by univariate Cox regression analysis (P=0.021), although this correlation was not statistically significant in the multivariate analysis (Table S1). When the MATH score was taken as a categorical variable, UCEC patients in the high-MATH group had a hazard of death that was twice as high as that of those in the low-MATH group [hazard ratio (HR), 2.342; 95% CI, 1.110–4.942; P=0.025; Table 2]. We also found that age, cancer type, clinical stage, histological grade, molecular type, and targeted molecular therapy were significantly associated with overall survival by univariate Cox regression (Table 2). To further investigate whether the MATH score was an independent prognostic factor, multivariate analysis was conducted with variables statistically significant in univariate analysis as covariates. The results showed that age (P=0.020) and clinical stage (P<0.001) were independent prognostic factors (Table 2). However, multivariate Cox regression showed that the MATH score, as a categorical variable, was not an independent prognostic factor in all patients (P=0.715; Table 2). Stratified analysis was then performed for age and clinical stage. Interestingly, the stratified analysis demonstrated that the MATH score was an independent prognostic factor that predicted the outcome of UCEC patients aged ≤60 years, when the MATH score was treated either as a
Full table
Full table
Full table
Full table
Full table
Full table
Full table
Full table
Evaluation of MATH for predictive ability in UCEC patients
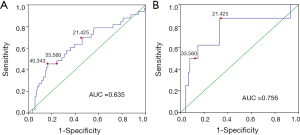
The diagnostic accuracy of the MATH score for predicting prognosis was assessed using the AUC. The diagnostic accuracy of the MATH score was higher in UCEC patients ≤60 years old (AUC, 0.756) than in all patients (AUC, 0.635) (Figure 7A,B). The predictive accuracy of different Cox models was described by the C-index. All variables in the predictive model, including age, cancer type, histological grade, clinical stage, molecular type, and targeted molecular therapy, were considered as known prognostic factors. For younger UCEC patients (age ≤60 years), the C-index of the predictive model was 0.699 (95% CI, 0.482–0.916), and the addition of MATH scores improved the C-index from 0.699 to 0.845 (95% CI, 0.749–0.941). However, there was no difference in C-index of the predictive model with or without MATH scores for all UCEC patients. These results indicate the MATH score is an additional prognostic variable for UCEC patients aged ≤60 years.
Discussion
Recent advances have demonstrated that UCEC is a complex disease characterized by a biologically, clinically, morphologically, and genetically heterogeneous group of tumors. Traditional classifications have not entirely taken this heterogeneity into account and have been limited in predicting responses to therapy, since heterogeneity is prognostic in nature. Despite numerous efforts to improve our understanding of the molecular mechanisms underlying UCEC, its clinical outcome in patients is not satisfactory and varies considerably due to the molecular heterogeneity of UCEC. In this study, we calculated the MATH scores to measure intra-tumor heterogeneity based on publicly available data obtained from TCGA, investigated their correlation with clinical characteristics, somatic mutations, or SCNA, and further explored the potential prognostic value of the MATH score in UCEC. We found that high MATH scores were significantly associated with poor clinical outcomes in UCEC patients, providing direct evidence—based on integrative genomic analysis—that high genetic heterogeneity, as indicated by high MATH scores, is related to short overall survival in UCEC patients.
A high MATH score indicates a high percentage of subclonal mutations, which are indicative of high intra-tumor heterogeneity, which also likely make the tumor more aggressive. Studies based on MATH scores have also been reported for other malignant human tumors. In HNSCC, high MATH scores were associated with advanced-stage tumors and shorter overall survival (12,13). Another study suggested that higher MATH scores in breast cancer were associated with tumor stage and triple-negative or basal-like subtypes (14,15). Moreover, high-MATH groups showed a tendency for worse overall survival in breast cancer patients (15). MATH was reported to be an independent risk factor in male colorectal cancer patients (16). Additionally, high MATH scores were correlated with higher risks of metastasis in stage-II and -III colon cancer (25). Recently, the MATH score has been shown to be a new parameter for describing the internal heterogeneity of lung tumors (17). Although the mechanisms underlying the links between high genetic heterogeneity and short overall survival cannot be determined from these data, our present results are consistent with the previous findings, confirming that MATH is a useful measure of intra-tumor heterogeneity and that it may serve as a novel biomarker for therapy selection and survival prediction for some human tumors.
A genomic classification of UCEC based on somatic mutations can entirely take into account this heterogeneity and identify potential targets for treatment in different subgroups of the disease (21). The current study found that TP53 was mutated significantly more often in patients with higher MATH scores at different clinical stages, mainly at advanced stages. Survival analysis indicated TP53 mutation was correlated with poor prognosis in UCEC patients. These results confirmed the findings of a previous study, which showed that TP53 was involved in the maintenance of genetic stability, and correlated with poor prognosis in breast cancer (26), lung cancer (27), and colorectal cancer (28). However, in this study, in both the subgroup with TP53 mutations and the TP53 wild-type subgroup, no significant difference in overall survival was found between the high- and low-MATH groups. Interestingly, in UCEC patients with PIK3CA mutations, overall survival was correlated with the MATH score. A similar tendency was found in UCEC patients with wild-type PTEN. These results suggest that the major influence of mutated PIK3CA or wild-type PTEN on the outcomes of UCEC patients may be a result of their tendency to regulate genetic heterogeneity, and even prognosis. Studies have demonstrated that quantification of whole-genome alteration, including the fraction of SCNA and somatic mutation count burden, can now be correlated with clinicopathologic characteristics and the therapeutic response, and may be useful in predicting patient outcomes (29,30). For example, a higher SCNA fraction was associated with advanced pathologic features, including tumor grade and stage, and was independently associated with poor survival in renal cell carcinoma cases from the TCGA database (31). Additionally, a high SCNA fraction was correlated with worse survival and platinum resistance in epithelial ovarian cancer (32). Notably, more SCNA events were independently related to the MATH score in male colorectal cancer patients, which might explain the gender-related prognostic value of the MATH score (16). Our present results demonstrated that the fraction of SCNA, which could contribute to the genetic heterogeneity measured by MATH, was indeed positively correlated with the MATH score in the TCGA UCEC cohort. Interestingly, Noorbakhsh et al. reported that MATH-based clinical predictions were driven by CN aberrations that altered mutant allele frequencies (33). Specifically, we found that the predictive power of the MATH score was robust in UCEC. This further supports the notion that measurement of heterogeneity by MATH was predictive of survival through an association with CN variation. It is yet to be determined whether SCNA can drive intra-tumor heterogeneity.
Our results showed that the UCEC patients ≤60 years old had significantly lower MATH scores than older patients, implying that the level of intra-tumor heterogeneity may be dependent on age. Previous reports have shown that the MATH score is similarly associated with adverse outcomes in clinically high-risk patients at advanced stages or with high histological grades, while the other groups have not been found to be associated with age (15,16,25). However, our present study revealed the MATH score is an independent prognostic factor for patients with UCEC who are ≤60 years old. Furthermore, the MATH score had better ROC characteristics, with an AUC above 0.7 in UCEC patients ≤60 years old, and was considered to provide higher predictive accuracy than the current prognostic factors, demonstrating it is a potential prognostic parameter to predict the outcome of UCEC in patients ≤60 years old. This could be explained in several ways. (I) Advanced age was considered to be a risk factor in endometrial carcinoma. However, this was not attributable to the fact that advanced-stage tumors are more likely to be found in older women, who tend to delay seeking medical care in the face of clinical symptoms. (II) Compared with older women, patients of ≤60 years old had an excellent survival and virtually no excess mortality during 5 years of follow-up. It was more helpful to objectively evaluate the prognostic value of different MATH scores. (III) The small number of patients in the cohort likely limited the multivariate analysis in this study.
Tumor heterogeneity is a potential cause of low efficacy or failure of therapies and may contribute to the development of drug resistance. As shown in Table 1, hormonal therapy resulted in a significant decrease in MATH scores, but the opposite results were found with respect to targeted molecular therapy; no significant differences in MATH score were found for radiation therapy. The findings of the present study indicate that individual treatment strategies may be adapted to tumor heterogeneity based on the MATH score, thereby improving the clinical outcomes of UCEC patients. The MATH score, in combination with clinical and pathological variables, can be used as a novel biomarker to improve our understanding of tumor heterogeneity, contribute to the identification of low-risk or high-risk tumors, and guide personalized treatment decisions.
Conclusions
Our findings suggest that high MATH scores were significantly associated with poor clinical outcomes among UCEC patients; the MATH score was an independent prognostic factor in UCEC patients ≤60 years old, and might therefore help to predict the survival of patients with UCEC. Our study had several limitations. Although much information about clinicopathologic parameters and outcomes was available from the TCGA database, these data were not collected prospectively, and many cases lacked long-term follow-up. The prognostic value of the MATH score needs to be further validated in additional independent cohorts with complete clinical information. Despite these limitations, our results improve our understanding of the quantification of intra-tumor heterogeneity, provide a potential prognostic biomarker for predicting UCEC outcomes, and contribute to the development of personalized therapeutic strategies in UCEC patients.
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of China (NSFC) Grant (No. 81773750, No. 81973325), and Beijing Natural Science Foundation Grant (No. 7194298).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethical approval was waived because we used only publicly available data and materials in this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015;27:15-26. [Crossref] [PubMed]
- Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 2018;15:81-94. [Crossref] [PubMed]
- Morris LG, Riaz N, Desrichard A, et al. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 2016;7:10051-63. [Crossref] [PubMed]
- Andor N, Graham TA, Jansen M, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 2016;22:105-13. [Crossref] [PubMed]
- Mota A, Colas E, Garcia-Sanz P, et al. Genetic analysis of uterine aspirates improves the diagnostic value and captures the intra-tumor heterogeneity of endometrial cancers. Mod Pathol 2017;30:134-45. [Crossref] [PubMed]
- Scheele CL, Maynard C, van Rheenen J. Intravital Insights into Heterogeneity, Metastasis, and Therapy Responses. Trends Cancer 2016;2:205-16. [Crossref] [PubMed]
- Joung JG, Oh BY, Hong HK, et al. Tumor Heterogeneity Predicts Metastatic Potential in Colorectal Cancer. Clin Cancer Res 2017;23:7209-16. [Crossref] [PubMed]
- McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017;168:613-28. [Crossref] [PubMed]
- Gara SK, Lack J, Zhang L, et al. Metastatic adrenocortical carcinoma displays higher mutation rate and tumor heterogeneity than primary tumors. Nat Commun 2018;9:4172. [Crossref] [PubMed]
- Andor N, Harness JV, Muller S, et al. EXPANDS: expanding ploidy and allele frequency on nested subpopulations. Bioinformatics 2014;30:50-60. [Crossref] [PubMed]
- Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol 2013;49:211-5. [Crossref] [PubMed]
- Mroz EA, Tward AD, Pickering CR, et al. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer 2013;119:3034-42. [Crossref] [PubMed]
- Mroz EA, Tward AD, Hammon RJ, et al. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med 2015;12:e1001786. [Crossref] [PubMed]
- Keenan T, Moy B, Mroz EA, et al. Comparison of the Genomic Landscape Between Primary Breast Cancer in African American Versus White Women and the Association of Racial Differences With Tumor Recurrence. J Clin Oncol 2015;33:3621-7. [Crossref] [PubMed]
- Ma D, Jiang YZ, Liu XY, et al. Clinical and molecular relevance of mutant-allele tumor heterogeneity in breast cancer. Breast Cancer Res Treat 2017;162:39-48. [Crossref] [PubMed]
- Zhang J, Yan S, Liu X, et al. Gender-related prognostic value and genomic pattern of intra-tumor heterogeneity in colorectal cancer. Carcinogenesis 2017;38:837-46. [Crossref] [PubMed]
- Mao H. Clinical relevance of mutant-allele tumor heterogeneity and lung adenocarcinoma. Ann Transl Med 2019;7:432. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Suarez AA, Felix AS, Cohn DE. Bokhman Redux: Endometrial cancer "types" in the 21st century. Gynecol Oncol 2017;144:243-9. [Crossref] [PubMed]
- Murali R, Soslow RA, Weigelt B. Classification of endometrial carcinoma: more than two types. Lancet Oncol 2014;15:e268-78. [Crossref] [PubMed]
- Piulats JM, Guerra E, Gil-Martin M, et al. Molecular approaches for classifying endometrial carcinoma. Gynecol Oncol 2017;145:200-7. [Crossref] [PubMed]
- Hussein YR, Soslow RA. Molecular insights into the classification of high-grade endometrial carcinoma. Pathology 2018;50:151-61. [Crossref] [PubMed]
- Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67-73. [Crossref] [PubMed]
- Mutation Analysis (MutSigCV v0.9) [database on the Internet]. Cambridge (Massachusetts): Broad Institute TCGA Genome Data Analysis Center. 2016. Accessed: www.firebrowse.org
- Rajput A, Bocklage T, Greenbaum A, et al. Mutant-Allele Tumor Heterogeneity Scores Correlate With Risk of Metastases in Colon Cancer. Clin Colorectal Cancer 2017;16:e165-70. [Crossref] [PubMed]
- Sheikh A, Hussain SA, Ghori Q, et al. The spectrum of genetic mutations in breast cancer. Asian Pac J Cancer Prev 2015;16:2177-85. [Crossref] [PubMed]
- Aisner DL, Sholl LM, Berry LD, et al. The Impact of Smoking and TP53 Mutations in Lung Adenocarcinoma Patients with Targetable Mutations-The Lung Cancer Mutation Consortium (LCMC2). Clin Cancer Res 2018;24:1038-47. [Crossref] [PubMed]
- Rodriguez-Salas N, Dominguez G, Barderas R, et al. Clinical relevance of colorectal cancer molecular subtypes. Crit Rev Oncol Hematol 2017;109:9-19. [Crossref] [PubMed]
- O'Sullivan DE, Johnson KC, Skinner L, et al. Epigenetic and genetic burden measures are associated with tumor characteristics in invasive breast carcinoma. Epigenetics 2016;11:344-53. [Crossref] [PubMed]
- Hieronymus H, Murali R, Tin A, et al. Tumor copy number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. Elife 2018. [Crossref] [PubMed]
- Ball MW, Gorin MA, Drake CG, et al. The Landscape of Whole-genome Alterations and Pathologic Features in Genitourinary Malignancies: An Analysis of the Cancer Genome Atlas. Eur Urol Focus 2017;3:584-9. [Crossref] [PubMed]
- Despierre E, Moisse M, Yesilyurt B, et al. Somatic copy number alterations predict response to platinum therapy in epithelial ovarian cancer. Gynecol Oncol 2014;135:415-22. [Crossref] [PubMed]
- Noorbakhsh J, Kim H, Namburi S, et al. Distribution-based measures of tumor heterogeneity are sensitive to mutation calling and lack strong clinical predictive power. Sci Rep 2018;8:11445. [Crossref] [PubMed]


