
Identification of TP53 mutation associated-immunotype and prediction of survival in patients with hepatocellular carcinoma
Introduction
Primary liver cancer, which consists predominantly of hepatocellular carcinoma (HCC), is the fifth most common type of cancer worldwide and the third most common cause of cancer mortality (1). Previously, considerable progress has been made in elucidating the molecular pathogenesis of HCC. HCC usually develops from chronic inflammatory liver disease (e.g., cirrhosis) and is associated with various pathogenic factors, such as hepatitis B virus (HBV), alcohol abuse, metabolic syndrome, hepatitis C virus (HCV), and diabetes. Only a few HCC patients are diagnosed at an early stage and are thus responsive to potentially curative treatments, such as locoregional procedures (radiofrequency ablation) and surgical therapies (resection and liver transplantation) (2). However, the HCC patients who are diagnosed with progression after locoregional therapy or those who are at an advanced stage with dismal prognosis, treatment with the front-line multi-tyrosine kinase inhibitor, sorafenib, and the second-line, regorafenib, may extend survival by 2 years (3).
In recent years, immune checkpoint inhibitors, which can activate the body’s self-immune response to attack tumors by blocking the “don’t eat me” label on T cells, have shown marked efficacy in different solid cancers, such as lung cancer or melanoma (e.g., ipilimumab, fresolimumab, pembrolizumab, and nivolumab) (4). Interestingly, although the checkpoint blockade strategy can lead to marked clinical responses, not all patients or cancer types have the same likelihood of responding to these regimens (5). Therefore, it is necessary and urgent to identify robust biomarkers that can effectively predict the therapy responses to checkpoint inhibitors and help in gaining an insight into the relationship between tumor subgroup and personalized treatment of HCC.
Tumor mutation burden (TMB) is a novel indicator for predicting the immune response in various types of cancer. Accumulating evidence indicates that tumors with high frequency of mutations are more likely to harbor neo-antigens, which makes them a target of activated immune cell attack (6). Mutations of TP53 and CTNNB1 genes are involved in the formation of intra-tumor heterogeneity and may contribute to treatment failure and drug resistance in many cases of HCC (7). However, mutations, especially TP53 mutations, play an important role in both tumor progression and therapeutic response.
Cytotoxic T lymphocyte (CTL), also known as CD8+ T cell, is thought to be related to the immunotherapy response. Previous studies suggest that the infiltration ratio of intratumoralCD8+ T cells represents the anticancer activity of the tumor, and that the tumors can be classified as “cold” and “hot”, based on the number of intratumoralCD8+ T cells (8). However, recent studies have found that only 10% of the intratumoral CD8+ T cells are able to recognize autologous tumors (9), while the remaining intratumoralCD8+ T cells may only be bystanders that lack anti-tumor reactivity (10).
Since neither of the two most important indicators is ideal, it is appropriate to characterize the immunotype subgroup of HCC according to the molecular features of intratumoral immune cells. This could provide more effective treatment strategies against special subtypes of HCC. In this study, we first observed that TP53 was the gene harboring the highest frequency of mutations in HCC patients, and TP53 mutations were immunogenic. Consequently, we aimed to identify TP53 mutation and immunocyte-associated lethal features, screened by least absolute shrinkage and selector operation (LASSO)-COX regression, and then establish TP53 mutation-associated immunotype subgroup of HCC, according to the lethal features. Moreover, various analyses have been performed to reveal the differences in prognosis, phenotype, and function between the two subgroups. Finally, a multivariate Cox analysis of overall survival (OS) was performed to evaluate the differences of hazard ratio (HR) between the two subgroups. An immunotype-based model and decision curve analysis (DCA) were established to evaluate the clinical value of immunotype in HCC patients.
Methods
Study design
As shown in Figure S1, this study was divided into three stages: screening of five lethal features, identification of TP53 mutation-associated immunotype, and the analysis of prognosis, phenotype, and function. First, single nucleotide polymorphism (SNP) analysis was performed for the top 30 genes with high mutation rate in 332 HCC patients, according to The Cancer Genome Atlas (TCGA) database, and for the top 20 genes with high mutation rate in HCC patients, according to the Catalogue of Somatic Mutations in Cancer (COSMIC) database. TP53 was confirmed to be the gene harboring the highest mutation frequency in HCC patients. Next, the data onTP53 mutations were integrated with 22 immune cells, which resulted in five lethal features, including TP53 mutations, according to LASSO-COX regression. Subsequently, 332 HCC patients (combined cohort) were randomly divided into training and validation cohorts to confirm the reliability of subsequent analysis. The patients in each cohort were divided into immunotype A and B subgroups. Finally, prognosis, phenotype, and functional analyses were carried out to explore the differences between the two subgroups, and the immunotype-based model and decision curve analysis (DCA) were established to evaluate the clinical value of immunotype in HCC patients (11).
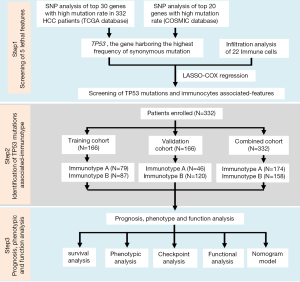
Patient population
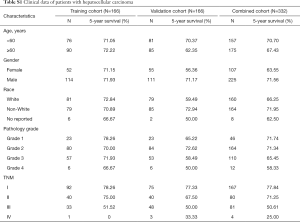
The mutation annotation format (MAF) document, mRNA expression data, and clinical data of HCC patients were downloaded from the publicly available TCGA data portal (up to June 13, 2018, https://tcga-data.nci.nih.gov). The following exclusion criteria were set: (I) patients were not diagnosed with HCC; (II) patients were diagnosed with HCC but died on the day of surgery; (III) patients had missing survival data, TNM stage, or other important clinicopathological features. Finally, as shown in Table S1, tumor samples from 332 patients were randomly divided into a training cohort and validation cohort, while the combined cohort consisted of all patients. This study abided by the publication guidelines provided by TCGA (12). The patient population of the COSMIC database was analyzed according to the default system.
Full table
Tumor mutational burden (TMB)
TMB was defined as the number of in-frame, missense, and truncating mutations, as well as deep deletion per megabase of the genome examined. The exome size was estimated as 38 Mb. The MAF files were downloaded from the official website of TCGA and used to determine TMB. The workflow type used for somatic mutation was the SomaticSniper pipeline (6). The mutation data of all genes, including TP53, were analyzed according to MAF files.
Analysis of infiltrating immune cells
The relative fractions of diverse cell subsets were accurately calculated by Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT) approach, according to the normalized mRNA expression data of 332 HCC patients (13). The following parameters were set: perm =100, QN =TRUE [perm is an integer number for the number of permutations, and QN is a Boolean value (TRUE or FALSE) for performing quantile]. Next, the fractions of infiltrating immune cells were integrated with TP53 mutation, TMB, and mRNA expression data of 332 HCC patients, according to patient ID and the corresponding clinical data.
Gene set enrichment analysis (GSEA)
The normalized expression data of all genes were acquired for conducting GSEA. GSEA, performed by MSigDB, was used to identify the pathways that were significantly enriched between the two immunotype subgroups, according to the selected signature. The number of random sample permutations was defined as 1,000. If a gene set had a positive enrichment score, then the set was termed “enriched”, which implied that the majority of the genes in the set had higher expression, accompanied by a higher risk score (14).
Gene set variation analysis (GSVA)
Pathway analyses were predominantly performed on the 50 hallmark pathways described in the molecular signature database by GSVA package. To reduce pathway overlaps and redundancies, each gene set associated with a pathway was trimmed to contain only unique genes, while all other genes associated with two or more pathways were removed. Most gene sets retained >70% of their associated genes (15).
Gene Ontology (GO) classification analysis
All differentially expressed genes, screened under conditions of |log2FC| >1 and FDR (adjusted P value) <0.05, were subjected to GO classification [GO-BP (biological process), GO-CC (cellular component), and GO-MF (molecular function)] to assign these genes to relevant GO Terms. R package org.Mm.eg.db was used to perform annotation and visualization (16). In particular, gene symbols of differentially expressed genes were converted to gene IDs, and the latter were subsequently used for functional enrichment analysis. Five immune-associated GO terms, with P values below 0.05, were visualized using bubble plots. Details of the genes belonging to particular GO terms with their fold change values were presented as circus plots using the “GOplot” library (17).
Expression analysis of mutated alleles and protein domain graphs
Expression of the main variants displayed in the oncoprints for the 332 HCC samples was determined and the frequency of the expressed allele was estimated. GenVisR R package was applied to calculate the mutation rate of genes and to construct the somatic mutation oncoprint (18). Protein domain graphs (PDGs) of the selected mutation genes were constructed using the Lollipop plot mutation diagram generator in maftools (19).
Pathway analysis
The network of immune-related signaling pathways was constructed with ConsensusPathDB, according to the gene lists that confirmed to be statistically significant between the two immunotype subgroups (20). A hypergeometric test was performed to analyze the over-represented pathways, based on the imputed gene list in the ConsensusPathDB website. The following pathway databases were selected for our analysis: Netpath, Humancyc, Inoh, Pid, Biocarta, Kegg, Reactome, Wikipathways, Ehmn, and Signalink, Smpdb, Pharmgkb. All the immune-related signaling pathways were selected and visualized according to the default settings: P value cutoff of 0.01 and minimum overlap with the input list of 2 (21).
Statistical analysis
Continuous variables were compared using the t-test or Wilcoxon rank-sum test, and the comparison of categorical variables was evaluated with the Chi-square test or Fisher’s exact test. The correlation analysis was carried out by Pearson correlation. OS was defined as the period from the date of surgery to the date of death (or the last follow-up). The Cox regression (proportional hazards) model was established to perform the univariate and multivariate analyses, and the characteristics with statistical difference in the univariate analysis were further assessed by the multivariate analysis. Survival curves were created using the Kaplan-Meier method, and the difference between groups was determined by the log-rank analysis. The accuracy of the prognostic model was evaluated by Harrell’s index of concordance (C-index). P<0.05 was considered statistically significant and all significance tests were two-sided (22).
The differential expression between the two subgroups was analyzed by “edgeR” package. The identification of up or downregulated genes was performed according to the following screening criteria: FDR (adjusted P value) <0.05 and at least 1-fold change. The median value was selected as the cutoff between the two groups, except in exceptional cases. The missing age of several patients was defaulted as 60 years. Nomogram and calibration plot, LASSO-COX regression, and cluster analysis were analyzed by R software package, version 3.4.2 (R Foundation for Statistical Computing, Vienna, Austria). Other statistical analyses were performed with Stata SE, version 13.0 (Stata, College Station, TX) and SPSS, version 20.0 (IBM, Armonk, NY).
Results
TP53 mutations suggested poor prognosis of HCC patients and were immunogenic
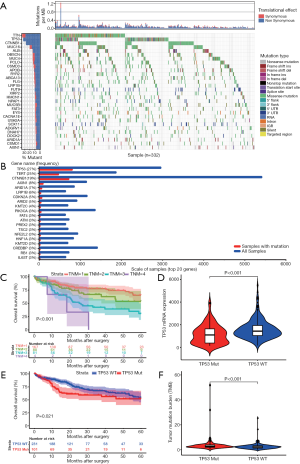
The mutational landscape of the top 30 genes in HCC, according to TCGA database, (Figure 1A) suggested that the TP53 gene harbored the highest frequency of non-synonymous mutation in HCC patients. The COSMIC database also showed the same result (Figure 1B). These results reiterated the fact that TP53 mutations play a dominant role in mutagenic mechanisms involved in cancer etiology. Next, the TNM stage was confirmed to be significantly associated with OS of HCC patients (Figure 1C, P<0.001). Subsequently, TP53 mutation was found to result in a lower expression level of TP53 mRNA (Figure 1D, P<0.001) and poorer OS of HCC patients (Figure 1E, P=0.021) when compared to TP53 wild type. TMB, a novel indicator used to predict the effectiveness of immunotherapy, was higher in TP53 mutation group compared to TP53 wild type group (Figure 1F, P<0.001), thus suggesting that there might be some associationbetweenTP53 mutation and immune response.
Screening of TP53 mutation and immunocyte-associated features and identification of immunotypes
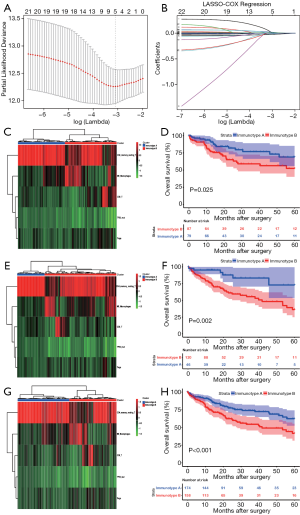
Since TP53 mutation was found to have an association with immune response, it was integrated with 22 immune cells for screening the lethal features according to LASSO-COX analysis. As shown in Figure 2, the minimum λ value resulted in five non-zero coefficients, viz., TP53 mutation, CD8+ T cells, CD4+ memory resting T cells (CD4+ T cells), regulatory T cells (Tregs), and M0 Macrophages (Macrophages). Subsequently, patients in the combined cohort were randomly divided into training (n=166) and validation cohorts (n=166) to confirm the reliability of subsequent analysis (Table S1). The three cohorts were all divided into two subgroups by clustering analysis, based on the five lethal features (Figure 2). An interesting observation in all the three cohorts was that the OS of HCC patients with immunotype B was consistently poorer compared to that in patients with immunotype A (P=0.025; P=0.002; P=0.001) (Figure 2).
Phenotypic analysis of immunotype subgroup
To understand the underlying cause of worse prognosis of HCC patients with immunotype B subgroup, we analyzed the proportion of five lethal parameters and carried out an enrichment analysis of immunocyte in the two subgroups. The infiltration ratio of CD4+ memory T cells was confirmed to be downregulated in immunotype B subgroup compared to that in immunotype A subgroup (Figure 3, P<0.001). In contrast, the infiltration rates of macrophages (Figure 3, P<0.001), Tregs (Figure 3, P<0.001), and CD8+ T cells (Figure 3, P<0.001) were clearly upregulated in immunotype B subgroup compared to that in immunotype A subgroup. These results were confirmed by GSEA analysis (Figure 3). The graphical summary of correlation among TP53 mutation, immunotype subgroups, and lethal immunocyte suggested that TP53 mutation-associated immunotype B subgroup resulted in lower infiltration of CD4+ memory T cells (Figure 3) and higher infiltration of macrophages (Figure 3), Tregs (Figure 3), and CD8+ T cells (Figure 3). It was consistent with the infiltration of CD8+ T cells that, the expression levels of interferon gamma (INFγ, Figure 3, P=0.004), granzyme B (GZMB, Figure 3, P=0.010) and perforin 1 (PRF1, Figure 3, P=0.014) mRNA were significantly upregulated in immunotype B subgroup compared with that in immunotype A subgroup although a worse prognosis in patients with immunotype B subgroup. Undoubtedly, these results changed our conventional wisdom about CD8+ T cells.
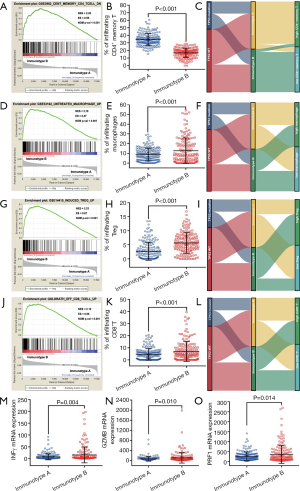
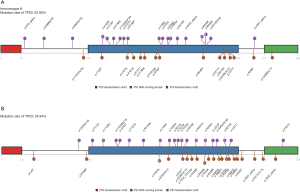
The location of mutations and protein domains of p53 (Figure 4) suggested that there were more mutations in the DNA-binding domain and tetramerization motif of p53 in immunotype B subgroup compared to immunotype A subgroup. The somatic mutation rate of TP53 was higher in immunotype B subgroup (34.94%) compared to that in immunotype A subgroup (25.90%).
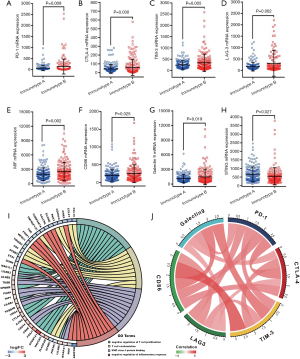
Immune checkpoints analysis between immunotype subgroups
To understand the underlying mechanism for the apparent contradiction between worse prognosis for HCC patients and higher infiltration of CD8+ T cells in the immunotype B subgroup, we compared the expression levels of the major immune checkpoints between the two immunotype subgroups. The expression levels of co-inhibitory immune checkpoints, namely PD-1 (programmed death-1, Figure 5, P=0.009), CTLA-4 (cytotoxic T-lymphocyte-associated protein 4, Figure 5, P=0.030), TIM-3 (T cell immunoglobulin domain and mucin domain-3, Figure 5, P=0.005), LAG-3 (Lymphocyte-activation gene 3, Figure 5, P=0.002), CD86 (Figure 5, P=0.025),and Galectin9 (Figure 5, P=0.019) were significantly upregulated in immunotype B subgroup compared to that in immunotype A subgroup. The expression level of the immune activator, STING (stimulator of interferon genes, Figure 5, P=0.027) was significantly downregulated, while the expression level of immunosuppression factor, MIF (macrophage migration inhibitory factor, Figure 5, P=0.002) was significantly upregulated in immunotype B subgroup compared to that in immunotype A subgroup. Circos plots of immunoreaction related GO terms and corresponding genes suggested that the expression of genes belonging to the terms “T cell costimulation” and “negative regulation of inflammatory response” were downregulated in immunotype B subgroup compared to that in immunotype A subgroup, while those belonging to the terms “MHC class I protein binding” and “negative regulation of T cell proliferation” were upregulated (Figure 5). Expectedly, the expression levels of immune checkpoints were highly correlated to each other (Figure 5).
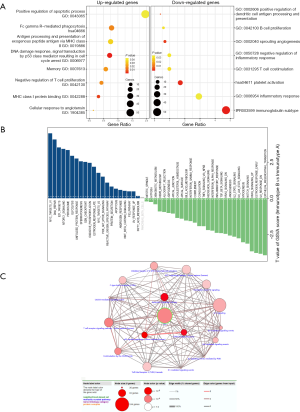
Enrichment of functions and signaling pathways in immunotype B subgroup
To investigate which functions and signaling pathways have changed in immunotype B subgroup compared to immunotype A subgroup, we carried out GO analysis and GSVA analysis. As shown in Figure 6A, functional enrichment analysis of differentially expressed genes in immunotype B vs. immunotype A suggested that the upregulated GO terms associated with immunity, according to upregulated genes (FDR <0.05, log2FC =1), mainly included positive regulation of DNA damage response, apoptotic process, and signal transduction by p53 class mediator, resulting in negative regulation of T cell proliferation, cell cycle arrest, and memory. Whereas, the downregulated GO terms associated with immunity mainly included T cell costimulation, platelet activation, inflammatory response, and immunoglobulin subtype. According to the expression of gene signatures in hallmark pathways, analysis of GSVA (Figure 6B) highlighted that most pathways were dramatically altered between immunotype B and immunotype A subgroups, which further confirmed that the two subgroups were completely different subsets. Myc targets were the top enriched signature in upregulated pathways, while the epithelial-mesenchymal transition was the top enriched signature in downregulated pathways. A network of immune-related signaling pathways was graphically presented, based on the differentially expressed genes between the two immunotype subgroups. As shown in Figure 6C, the signaling pathways associated with inflammation, immune responses, or DNA damage response were found to play a dominant role in the network.
Multivariate Cox analysis of overall survival and nomogram prediction of clinical outcomes for patients with hepatocellular carcinoma
To further analyze the possibility of immunotype as a potential independent prognostic factor for HCC patients, univariate and multivariate Cox analyses were successively performed. As shown in Table S2, immunotype was found to be negatively associated with OS, according to univariate Cox analysis (P<0.001). Additionally, the TNM stage was significantly associated with OS of HCC patients. In contrast, age, gender, race, and pathology grade were not correlated with OS. Subsequently, immunotype and TNM stage were incorporated into the multivariate Cox analysis for the three cohorts (Figure 7A). Immunotype and TNM stage were confirmed to be independent hazard markers for OS in the validation cohort and combined cohort. Although there was no significant association between immunotype and OS, immunotype B subgroup was also confirmed to be a risk factor in the training cohort.

Full table
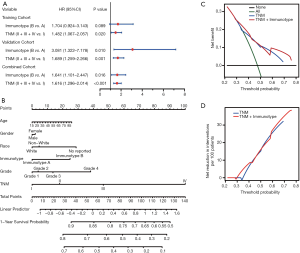
A newly built nomogram model via integration of the traditional risk factors mentioned above, including immunotype, was established to predict postoperative survival of HCC patients, and named as “immunotype-based model”. As demonstrated in Figure 7B, total points were used to assess the cumulative risk level of every risk factor. The higher the total point, the shorter was the OS of HCC patients. The predictive accuracies of the immunotype-based model and TNM stage were evaluated by C-index (Harrell’s concordance index). As shown in Table S3, the C-index of TNM stage for 3-year OS was 0.614, and the C-index of the immunotype-based model was 0.658. Similarly, the C-index of TNM stage for 5-year OS was 0.613, while the C-index of the immunotype-based model for 5-year OS was 0.656. These results indicated that the predictive accuracy of the immunotype-based model was better than that of TNM stage for OS of HCC patients.

Full table
A net decision curve was used to perform the OS predictions (Figure 7C). Compared to TNM stage alone, the combined analysis of TNM stage and immunotype clearly benefited those patients with a survival probability of less than 42% or more than 60%. For instance, if the survival probability of 35% was used as a threshold, the net benefit of the combined model was approximately 0.23, which was superior to 0.19 for TNM stage. Similarly, the net reduction analyses (Figure 7D) demonstrated that the model based on TNM stage and immunotype could avoid more interventions of survival prediction for those patients with a survival probability of about less than 42% or more than 60%. Therefore, the immunotype-based model was not only accurate but also had practical superiority compared to the TNM stage.
Discussion
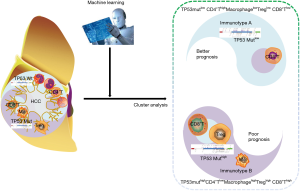
In this study, it was shown that the TP53gene harbored the highest frequency of mutations and these mutations were immunogenic in HCC patients. Subsequently, five lethal features, including TP53 mutations, were screened by LASSO-COX regression according to TP53 mutations and 22 infiltrating immune cells. Consequently, two distinct subgroups of HCC were identified, based on the previous five lethal features. TP53 mutation-associated immunotype B subgroup had poor prognosis, lower infiltration of CD4+ memory cells and higher infiltration of macrophage, Tregs and CD8+ T cells as well as a higher rate of TP53 mutations. Furthermore, the expression levels of co-inhibitory immune checkpoints were significantly upregulated, while the GO terms or pathways to boost immune responses were inhibited in immunotype B subgroup compared to that in immunotype A subgroup. Finally, the immunotype was proved to be an independent adverse prognostic factor contributing to improvement in predictive accuracy of the immunotype-based model and avoiding excessive medical treatment of some HCC patients (Figure 8).
TP53 mutations are one of the most frequent alterations in human cancers. These mutations have been integrated into the International Agency for Research on Cancer TP53 database (http://www-p53.iarc.fr/) (23). Interestingly, single-base substitutions distributed throughout the coding sequence are the most common form of TP53mutation, however, diverse types and positions of the mutations may represent diverse mutagenic mechanisms involved in cancer etiology (24). Mutations in the TP53 gene can give rise to genetic instability and lead to cancer progression. This is because TP53 mutations abrogate the functions of the p53 protein, such as gene transcription, DNA synthesis and repair, cell cycle arrest, senescence, and apoptosis, and the key responder to oxidative and nitrosative stress (25). Recent accumulating evidence suggests thatTP53mutations are immunogenic in epithelial cancer (26). TP53 mutation significantly increases the expression of immune checkpoints, activates effector T cells and interferon-γ signature, and may serve as a potential predictive factor in guiding anti-PD-1/PD-L1 immunotherapy in lung adenocarcinoma (27). The miR-1246-enriched exosomes, shed by colon cancer cells harboring TP53 mutants, are uptaken by neighboring macrophages which then triggersmiR-1246-dependent reprogramming into a cancer-promoting state and favors anti-inflammatory immunosuppression with increased activity of TGF-β (28). Herein, we found that TP53 mutants suggested higher mutation burden of HCC patients (Figure 1F) and more enrichment of the immunotype B subtype, named as TP53 mutation-associated immunotype B subtype (Figure 2C,E,G; Figure 4B). Moreover, TP53 mutation-associated immunotype B subtype resulted in more infiltration of Treg, macrophage, and CD8+ T cells (Figure 3F,I,L) as well as more adverse prognosis (Figure 2D,F,H). These results were consistent with findings from previous studies.
The stratification of tumors is paramount to achieve better clinical outcomes. Inactivation of CDK12was reported to result in a distinct immunological phenotype in patients with advanced prostate cancer (29). The mesenchymal-like type, including diffuse-subtype tumors with the worst prognosis, has the highest recurrence frequency (63%) among the four subtypes, based on disease progression, molecular alterations, and prognosis of patients with gastric cancer, and tends to occur at an earlier age (30). A novel genomic subtype with TP53 and SETDB1 mutations and extensive loss of heterozygosity and strong expression of the immune-checkpoint gene VISTA, is defined according to the comprehensive integrated genomic study of 74 malignant pleural mesotheliomas (MPMs) (31). However, although at a cursory glance, the complexity of classification might be increased because of this subdivision of organ-specific cancers, further analysis suggests that the subtypes detected in the various malignancies are recurring. For example, nearly all subtypes of gastrointestinal cancers are characterized with extensive immune infiltration, mesenchymal gene expression signatures, or metabolic dysregulation (32). In this study, we identified two distinct immune subtypes with different prognosis (Figure 2), different proportion of immune cells (Figure 3), different probability of TP53 mutations (Figure 4), and different functions (Figures 5,6). These results may contribute in providing different therapeutic strategies for the two distinct immune subtypes. It has been widely reported that higher the proportion of intratumoralCD8+T infiltration, the better is the prognosis of the patients. However, in our study we obtained contradictory results. The infiltration ratio of CD8+T cells was significantly higher in immunotype B subgroup than that in immunotype A subgroup (Figure 3). Yet, the patients in immunotype B subgroup had a shorter survival compared to patients in immunotype A subgroup (Figure 2). This observation subverted the traditional understanding of intratumoral CD8+ T cells. The reasons underlying this phenomenon may lie in immunotype of the tumor and should be determined by multiple omics analyses rather than solely determining the infiltration proportion of immune cells (32,33). Further, the CD8+ T cells may have many different subsets, such as naive, exhausted, and effector CD8+ T cells (34).
The most promising approach to activate therapeutic antitumor immunity is the blockade of immune checkpoints (35). Overexpressing the ligands of checkpoint receptors on tumor cells or adjacent cells results in T-cell exhaustion and immune tolerance (36). Accumulating evidence suggests that blockade of cytotoxic T-lymphocyte-associated antigen 4 (CTLA4) or programmed cell death-1 (PD-1) pathway significantly inhibits tumor growth and improves the survival of patients (37). However, differences have been observed with respect to patient’s response to PD-1/PD-L1 blockade treatment strategy. While some patients have achieved significant results, for others, this strategy has proved to be ineffective (38). PD-1/PD-L1 immunotherapy is ineffective in some cancer patients, and the most important reason for this is the presence of a large number of “Bystander” CD8+ T cells (about 90%) without killing capability in human tumor tissues (9,10). In this study, we found that although the infiltration ratio of intratumoral CD8+ T cells (Figure 3J,K,L) and the expression levels of cytotoxic effector factors (Figure 3M,N,O) were significantly higher in immunotype B subgroup than that in immunotype A subgroup. Expectedly, the expression levels of immune checkpoints (Figure 5) were significantly upregulated and the functions and pathways associated with immune response (Figure 6) were evidently inhibited in the immunotype B subgroup. Moreover, the OS of HCC patients (Figure 2) was significantly shorter in immunotype B subgroup compared to immunotype A subgroup. Therefore, these results suggest that tumors should not be simply distinguished as “cold” and “hot” based on the number of infiltrating CD8+T cells in the tumor. More importantly, it is important to estimate whether the TCR of infiltrating CD8+T cells can recognize the tumor.
The molecular subtype of HER2 and ER, as a stronger predictor, contributes to building a nomogram to calculate the probability of sentinel node positivity in breast carcinoma (39). In this study, immunotype and TNM stage were all found to be clearly associated with OS of HCC patients, based on the results from the univariate Cox analysis (Table S2), and were also proved to be independent adverse prognostic factors for OS (Figure 7A). The immunotype-based model was not only accurate but also had practical superiority compared to the TNM stage (Figure 7B,C,D, Table S3). Our data suggested that TP53 mutation-associated immunotype could serve as an independent risk factor and contribute to improvement of predictive accuracy and avoid excessive medical treatment of some HCC patients. It is important to note that several conventional factors were incorporated into the nomogram model, although there was no significant association between them and OS.
Although this study successfully analyzed the prognosis, phenotype, and function of TP53 mutation-associated immunotype for HCC patients, it had a few limitations. In future, the molecular mechanisms of immune evasion in TP53 mutation-associated immunotype B subgroup of HCC patients should be confirmed further through several basic experiments. Additionally, a more detailed subgroup clustering should be performed such that the curve of the immunotype-based model is plotted absolutely above the curve of TNM stage. Further, a well-designed, multicenter clinical trial is required to validate the independent prognostic value of immunotype-based nomogram model and potential guiding significance for adjuvant chemotherapy and immunotherapy. Clearly, HCC etiology, such as Hepatitis B virus (HBV), hepatitis C virus (HCV), alcohol consumption, and diabetes mellitus are important factors affecting the divide-and-conquer strategy of HCC (40). For example, anti-HBV treatment should be consistently carried out for patients with HBV-associated HCC, even after radical resection of HCC. Although we did not incorporate it into our study due to incomplete or missing clinical data, in the future, it should be incorporated into the subgroup analysis as far as possible.
In summary, we identified two distinct subgroups of HCC in terms of prognosis, phenotype, and function. The immunotype contributed to improving the predictive accuracy of the immunotype-based model for HCC patients and avoiding excessive medical treatment of those patients with a survival probability of less than 42%, or more than 60%. Therefore, TP53 mutation-associated immunotype is paramount to achieve better clinical outcomes and might have crucial implications for the postoperative personalized follow-up and decision-making during further individualized treatment.
Acknowledgments
The authors gratefully acknowledge contributions from the TCGA Network.
Funding: This study was funded by grants from the National Natural Science Foundation of China (No. 81401988), China Postdoctoral Science Foundation (2019M661907), Jiangsu Postdoctoral Science Foundation (2019K159, 2019Z153), General Project of Jiangsu Provincial Health Committee (H2019101), the Six Talent Peaks Project in Jiangsu Province (WSN-060) and the Key Talents of Youth Medicine in Jiangsu Provincial Department of health (QNRC2016681). All these study sponsors have no roles in the study design, in the collection, analysis, and interpretation of data.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study had been approved by the Ethics review committee, Affiliated Hospital of Nantong University (2017-K025). Consent for participation of all patients was obtained through The Cancer Genome Atlas Project.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Yi SW, Choi JS. Reply to Risk factors for hepatocellular carcinoma. Cancer 2019;125:483. [Crossref] [PubMed]
- Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378-90. [Crossref] [PubMed]
- Sia D, Jiao Y, Martinez-Quetglas I, et al. Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology 2017;153:812-26. [Crossref] [PubMed]
- Zheng C, Zheng L, Yoo JK, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017;169:1342-1356.e16. [Crossref] [PubMed]
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [Crossref] [PubMed]
- Friemel J, Rechsteiner M, Frick L, et al. Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res 2015;21:1951-61. [Crossref] [PubMed]
- Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321-30. [Crossref] [PubMed]
- Scheper W, Kelderman S, Fanchi LF, et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat Med 2019;25:89-94. [Crossref] [PubMed]
- Simoni Y, Becht E, Fehlings M, et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 2018;557:575-9. [Crossref] [PubMed]
- Qiu J, Peng B, Tang Y, et al. CpG Methylation Signature Predicts Recurrence in Early-Stage Hepatocellular Carcinoma: Results From a Multicenter Study. J Clin Oncol 2017;35:734-42. [Crossref] [PubMed]
- Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity 2018;48:812-830.e14. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Yoon S, Nguyen HCT, Yoo YJ, et al. Efficient pathway enrichment and network analysis of GWAS summary data using GSA-SNP2. Nucleic Acids Res 2018;46:e60. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:3. [Crossref] [PubMed]
- Walter W, Sanchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 2015;31:2912-4. [Crossref] [PubMed]
- Skidmore ZL, Wagner AH, Lesurf R, et al. GenVisR: Genomic Visualizations in R. Bioinformatics 2016;32:3012-4. [Crossref] [PubMed]
- Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med 2016;375:443-53. [Crossref] [PubMed]
- Kamburov A, Wierling C, Lehrach H, et al. ConsensusPathDB--a database for integrating human functional interaction networks. Nucleic Acids Res 2009;37:D623-8. [Crossref] [PubMed]
- Kamburov A, Stelzl U, Lehrach H, et al. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res 2013;41:D793-800. [Crossref] [PubMed]
- Fu H, Zhu Y, Wang Y, et al. Identification and Validation of Stromal Immunotype Predict Survival and Benefit from Adjuvant Chemotherapy in Patients with Muscle-Invasive Bladder Cancer. Clin Cancer Res 2018;24:3069-78. [Crossref] [PubMed]
- Leroy B, Anderson M, Soussi T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat 2014;35:672-88. [Crossref] [PubMed]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2010;2:a001008. [Crossref] [PubMed]
- Stracquadanio G, Wang X, Wallace MD, et al. The importance of p53 pathway genetics in inherited and somatic cancer genomes. Nat Rev Cancer 2016;16:251-65. [Crossref] [PubMed]
- Malekzadeh P, Pasetto A, Robbins PF, et al. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J Clin Invest 2019;129:1109-14. [Crossref] [PubMed]
- Dong ZY, Zhong WZ, Zhang XC, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res 2017;23:3012-24. [Crossref] [PubMed]
- Cooks T, Pateras IS, Jenkins LM, et al. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun 2018;9:771. [Crossref] [PubMed]
- Wu YM, Cieslik M, Lonigro RJ, et al. Inactivation of CDK12 Delineates a Distinct Immunogenic Class of Advanced Prostate Cancer. Cell 2018;173:1770-1782.e14. [Crossref] [PubMed]
- Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449-56. [Crossref] [PubMed]
- Hmeljak J, Sanchez-Vega F, Hoadley KA, et al. Integrative Molecular Characterization of Malignant Pleural Mesothelioma. Cancer Discov 2018;8:1548-65. [Crossref] [PubMed]
- Bijlsma MF, Sadanandam A, Tan P, et al. Molecular subtypes in cancers of the gastrointestinal tract. Nat Rev Gastroenterol Hepatol 2017;14:333-42. [Crossref] [PubMed]
- Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017;171:540-556.e25. [PubMed]
- Guo X, Zhang Y, Zheng L, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med 2018;24:978-85. [Crossref] [PubMed]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252-64. [Crossref] [PubMed]
- Wang Y, Wu L, Tian C, et al. PD-1-PD-L1 immune-checkpoint blockade in malignant lymphomas. Ann Hematol 2018;97:229-37. [Crossref] [PubMed]
- Ott PA, Hodi FS, Robert C. CTLA-4 and PD-1/PD-L1 blockade: new immunotherapeutic modalities with durable clinical benefit in melanoma patients. Clin Cancer Res 2013;19:5300-9. [Crossref] [PubMed]
- Liang Y, Tang H, Guo J, et al. Targeting IFNalpha to tumor by anti-PD-L1 creates feedforward antitumor responses to overcome checkpoint blockade resistance. Nat Commun 2018;9:4586. [Crossref] [PubMed]
- Reyal F, Rouzier R, Depont-Hazelzet B, et al. The molecular subtype classification is a determinant of sentinel node positivity in early breast carcinoma. PLoS One 2011;6:e20297. [Crossref] [PubMed]
- Chitapanarux T, Phornphutkul K. Risk Factors for the Development of Hepatocellular Carcinoma in Thailand. J Clin Transl Hepatol 2015;3:182-8. [Crossref] [PubMed]


