
The implementation of drug reposition for alcoholic hepatitis based on a sub-pathway integration strategy
Introduction
The most serious form of alcoholic liver disease (ALD) is alcoholic hepatitis (AH), most commonly called AH (1). It is a clinical syndrome that is usually characterized by damage to hepatocellular tissues, steatosis, and pericellular fibrosis (2). Typically, AH is associated with alcohol misuse and usually exhibits progressive jaundice and inflammatory liver injuries (2,3). Severe cases have poor short-term prognosis, and if observed for 28 days, a death rate of around 30–50% (4,5). In addition to this, there is yet no definitive pathological process responsible and thus a lack of developed targeted therapy (6).
Considerable progress has been made toward the characterization of immune response for this disease. These responses include the development of both pro- and anti-inflammatory cytokines in the blood, trafficking of immune cells to the liver, and determining the mechanism of cell death (3,7-9). However, the unavailability of animal models for studying AH has limited further progression in these types of studies (5). Recent research has shown that the drugs targeting inflammatory cytokines such as tumor necrosis factor (TNF) may play an essential role in AH, and thus, drugs interfering with TNF were tested in patients (8-10). Unfortunately, the results observed from these studies were disappointing as they led to an increased risk of fatal bacterial infections in patients treated with TNF (11,12). Therefore, targeted strategies for the treatment of AH are still unavailable.
It is known that the de novo development of drugs is time-consuming, costly, and has no guarantee for success. Meanwhile, another innovative approach, referred to as drug repositioning, has been developed due to advances in computational biology and the availability of public databases (13). The Gene Expression Omnibus (GEO) and the connectivity map (CMap) databases provide considerable microarray datasets which were obtained either under diseased conditions or under the effects of drugs (14). In this study, a pathway-based method responsible for the drug repositioning of AH was conducted by using a bioinformatics analysis, which was followed by experimental validation. Initially, the metabolic mechanisms of AH were explored, which was followed by the identification of potential small molecular drugs. The drugs were selected in such a way that they could target metabolic sub-pathways which undergo dysregulation during the disease’s development and progression. Finally, via in vitro study, our data indicated that the treatment of riboflavin, one of the agents we identified, could reverse ethanol-induced AML-12 cell injury. In brief, our research may offer significant insight into the potentially effective therapies for AH.
Methods
Identification of gene expression using microarray data
Microarray data for GSE28619 were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) provided by the National Center of Biotechnology Information (NCBI). In this profile, hepatic gene expression was assessed in patients with AH (n=15) and controls with normal livers (n=7) using the Affymetrix Human Genome U133 Plus 2.0 Array platform (6). The data obtained from microarray experiments and the probe annotation files were then downloaded for the analysis. Here, 2 methods were used for the identification of genes undergoing differential expression, commonly known as DEGs (differentially expressed genes), and the overlapping gene set was considered as the final DEGs of interest. At first, if 1 gene matched more than 1 probe, the expression value of this gene was computed by taking the average expression value of all the corresponding probes. We then converted each probe into a gene by using the Entrez Gene IDs, and the fold change was calculated for all the genes. The genes that showed a higher than 2 or lower than 0.5-fold change was selected. A total of 1,123 genes were obtained. Next, we used the SAM method to identify DEGs, and genes with a q-value <0.01 were selected (4,337 genes were obtained). Finally, the overlapping genes between the 2 sets of data (1,040 genes) were considered as DEGs.
Identification of AH-related metabolic sub-pathways
SubpathwayMiner R package, which is a pathway identification method known to be flexible for sub-pathway determination was used (15) to determine which sub-pathways were perturbed during AH. This software firstly converted the pathway structure data present in the Kyoto Encyclopedia of Genes and Genomes (KEGG) to undirected R graph objects. Thereafter, the enzymes were regarded as nodes. The 2 nodes which belonged to a single reaction were connected by an edge in an undirected graph. Finally, SubpathwayMiner could divide the entire pathway into sub-pathways using the “k-clique” method, which is defined as a sub-graph in which the distance between any 2 nodes is no greater than a parameter, k, in social network analysis (15). Here, k=3 was the parameter set, which suggests that the gap between the enzymes in a single sub-pathway was lower than 3.
After our DEGs were put into SubpathwayMiner, the results obtained indicated the significantly enriched sub-pathways of AH. A P value <0.01 was considered as the cutoff criterion for statistically significant metabolic sub-pathways.
The connections between potential drug targets and sub-pathways involved in metabolism
For the identification of the novel small molecular drugs that might affect important sub-pathways involved in AH, we considered the data on the global associations which were observed between drugs and the metabolic sub-pathways provided by Li and coworkers (16). Li and colleagues analyzed microarray data from cells that were pre-treated with small molecular drugs determined from the Cmap database and thus identified the DEG set for every drug using a cutoff with a |log(fold change)|>1 (16). Then, the sub-pathways which were affected by drugs were identified using the SubpathwayMiner software package (P value <0.01) (16). Subsequently, a total of 3,925 associations were obtained between the drugs and the metabolic sub-pathways, that were affected, comprising 488 drugs and 403 metabolic sub-pathways. Out of the obtained 403 sub-pathways, 6 sub-pathways (corresponding to 64 drugs) were hampered by AH as well.
Cell culture
AML-12 cells were used for this study, and these cells were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in F12 medium in the presence of 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 µg/mL streptomycin. The culture medium and the antibiotics were purchased from Invitrogen (USA) while the FBS was purchased from Invitrogen (Carlsbad, CA, USA). The cells used for the study were grown and kept under humidified conditions in the presence of 5% CO2 at a temperature of 37 °C.
Cell Counting Kit-8 (CCK-8) cell viability assay
AML-12 cells were treated with different concentrations of ethanol (0, 50, 75, 100, 150, 200) mM or riboflavin (Sigma, USA) at the concentrations of 30 or 60 nM. Cells were seeded in 96-well plates at a density of 2×103 per well and cultured for 24 h. Cell viability was assessed by the Cell Counting Kit-8 (CCK-8, Dojindo, Japan).
Biochemical assays
The levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were determined using commercial kits that were procured from Nanjing Jiancheng Bioengineering Institute located in Nanjing, China. The amount of ethanol in the blood was then determined by a kit which was procured from BioVision Inc. (San Francisco, CA, USA). The experiments using the procured kits were performed as per the manufacturer’s instructions.
Statistical analysis
The data obtained from these experiments were analyzed using SPSS 19.0 (Chicago, IL, USA). The results of the experiments are stated as the mean values, along with the standard deviation (SD) obtained from the multiple sets of experiments. A comparison between multiple groups was conducted by use of the analysis of variance (ANOVA) and Student-Newman-Keuls method. Differences were statistically significant at a P value <0.05.
Results
Analysis of DEGs between AH and healthy controls
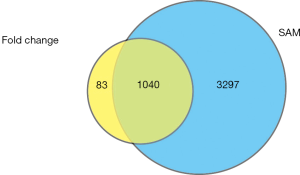
We obtained the mRNA expression profiles of AH samples and normal control samples from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The GSE28619 dataset, which was also available in the GEO database, was downloaded, and then the DEGs between AH patients and controls were identified by fold changes and the SAM method (6). A total of 1,041 genes were determined to be expressed differentially (Figure 1; fold change >2 or <0.5 and q-value <0.01; see Methods).
Exploration of the AH mechanism based on sub-pathway enrichment analysis
For the exploration of the metabolic mechanisms of AH, the Subpathwayminer was used to identify AH-involved metabolic sub-pathways (15). After annotating the DEGs onto the metabolic sub-pathways (k=3), 11 enriched metabolic sub-pathways were identified which corresponded to 8 total metabolic pathways having a strict cutoff P value (P value <0.01) as shown in Table 1. As a result of these experiments, 11 metabolic sub-pathways were identified as being related to AH development.

Full table
AH drug repositions
Next, we identified candidate agents that could potentially target AH-related metabolic sub-pathways, using the dataset of global associations observed between the drugs and the metabolic sub-pathways published by Li and colleagues (16) (see the Methods).
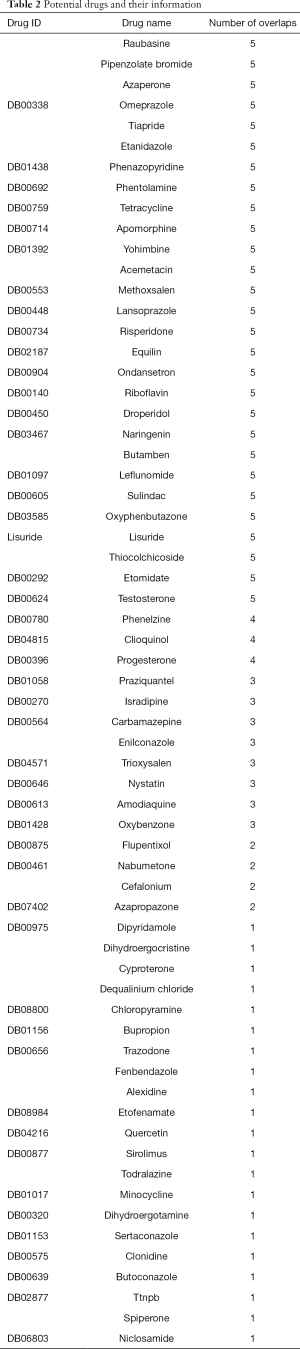
After the integration of the 11 AH-related sub-pathways and the 403 sub-pathways affected by drugs, the overlapping sub-pathways, which were both related to AH and affected by small molecular drugs, were obtained. The drug molecules which could affect the sub-pathway are potential therapeutic candidates to target the related pathways of AH. Sixty-four small molecules were identified in total (Table 2).
Full table
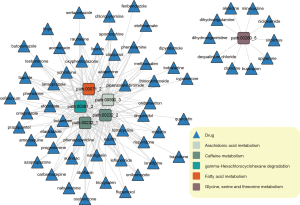
This was followed by the building of a bipartite network as per the predicted drugs and the overlapping metabolic sub-pathways by the integration of the above relationships, as stated in Figure 2. We found that some drugs could perturb up to 5 metabolic sub-pathways which corresponded to 4 complete pathways (fatty acid metabolism, caffeine metabolism, gamma-hexachlorocyclohexane degradation, caffeine metabolism, and arachidonic acid metabolism), whereas others could only perturb 1 sub-pathway (glycine, serine, and threonine metabolism; right part of Figure 2).
Potential pathway of riboflavin in the metabolism of purines
For the prediction of the potential mode of action of riboflavin in AH, the DEGs for AH and the DEGs for riboflavin were mapped to the pathway responsible for the metabolism of purines (path: 00230) by using the “user data mapping” tool which is available in the KEGG database (http://www.genome.jp/kegg/) (Figure 3). The DEGs of riboflavin were obtained from the supplementary datasets, which were provided by Li and colleagues (16). The DEGs of riboflavin and AH are indicated by blue and red rectangles, respectively, in Figure 3.

Protective effects of riboflavin on alcoholic liver injury
The effect of riboflavin was determined in liver injury which is induced by alcohol usage. We first explored whether treatment with alcohol can induce AML-12 cell injury. We evaluated the cell viability of alcohol at different concentrations (0, 50, 75, 100, 150, 200) mM for 24 h. As observed in Figure 4A, the cell viability of the used cells was found to decrease in a dose-dependent manner. We also determined the levels of AST and ALT, which was the standard of liver injury. Our experiments also showed that the level of AST or ALT was decreased in a dose-dependent manner (Figure 4B,C). This was followed by stimulation of AML-12 cells with 100 mM EtOH for 24 h, which could serve as an alcoholic in vitro model for the study.

To identify the regulatory effect of riboflavin towards ethanol-induced AML-12 injury, we detected the cell viability of AML-12 treated with riboflavin (0, 10, 30, 60, 100) nM for 24 h. As displayed in Figure 4B,C, the results show that, compared to the control group, the levels of ALT and AST, and the activity of these enzymes were higher in the alcohol group. However, compared to ethanol alone, riboflavin treatment significantly lowered ALT and AST activity in a dose-dependent manner, thereby suggesting that riboflavin shows a protective effect against liver injury caused by alcohol. Collectively, the results from these experiments indicate that riboflavin is an effective molecule in protecting ethanol-induced liver cell injury.
Discussion
Microarray analysis is a high-throughput screening technology that is considered an essential tool for analyzing gene expression patterns and providing possible targets for improvements in therapeutic interventions. In the present study, microarray data of laparoscopic liver biopsy specimens of AH patients were used to explore the basic mechanisms for the development of AH. We used strict criteria for the identification of DEGs; i.e., genes comprising of a fold change >2 or <0.5 and a q-value <0.01 using the SAM method. As a result, 1,040 DEGs were identified. With the use of the SubpathwayMiner software, it was indicated that there were 11 metabolic sub-pathways, which corresponded to a total of 8 pathways, and were dysregulated during AH. The potential small molecules for AH were then predicted by the integration of these sub-pathways that were related to AH and the drugs used. Finally, a total of 64 small molecules that might perturb the development of AH were identified.
Unlike other methods, the sub-pathway approach focuses on a certain local area of the entire pathway, suggesting that it can identify more subtle changes that are always neglected by the analysis of complete pathways. This may, therefore, be a more suitable and more flexible approach for understanding drug responses (15,16). In the present study, 11 sub-pathways, which corresponded to 8 complete KEGG metabolic pathways, were identified. A few of these sub-pathways have been reported to play important roles in AH (Table 1). For example, the most significant sub-pathway detected was shown to be involved in fatty acid metabolism (path:00071_2; P value =2.97-16). It is known that the accumulation of fat in the form of lipid droplets within hepatocytes can result in hepatic steatosis, which is a hallmark of liver diseases like AH (17,18). Furthermore, dietary fatty acids and obesity have been shown to affect the degree of fat accumulation within the liver (17,18). Therefore, fatty acid metabolism appears to play a key role in the development of AH. The “caffeine metabolism” pathway (path: 00232) was also identified to be among the affected sub-pathways. Studies have indicated that caffeine clearance is enhanced during AH compared with controls (19). The “arachidonic acid metabolism” (path: 00590) path was also found to be broadly distributed in AH, suggesting there to be abnormal microsomal elongation activity in the liver cells (20).
We also identified a group of drugs with possible therapeutic efficacy for the disease. The findings of the studies in the current study culminated in 64 small molecules being identified. Some molecules share affected sub-pathways with AH, and it is understood that some of these agents are active against AH (testosterone) and other potentially new agents. Testosterone, a steroid sex hormone, has the ability to robustly inhibit hepcidin transcription associated with the development of liver disorders such as liver disease (21).
Decades ago, testosterone was shown to be efficacious in the removal of hepatic fat, which is variably present among patients with acute AH (22). Riboflavin, also known as vitamin B2, is an easily absorbed, water-soluble micronutrient with a critical role in maintaining human health. A deficiency of riboflavin has been reported among patients with a histologically normal liver but not among other groups (23). However, there have been no direct reports of the therapeutic management of riboflavin in AH patients, and the underlying mechanisms are still unclear. To investigate the potential mechanisms of action of riboflavin on AH, we annotated the DEGs of AH and riboflavin in the most significant pathway “fatty acid metabolism” (Figure 3). Annotation of the DEGs for AH identified alcohol-metabolizing enzymes, including aldehyde dehydrogenase (ALDH) and alcohol dehydrogenase (ADH), whose genetic mutation was associated with the development of acute AH (24).
Furthermore, the DEGs for riboflavin included CYP1B1 and CYP1A1, which participate in the same or consecutive reactions with ADH and ALDH. Our data showed that riboflavin treatment could reverse ethanol-induce AH and markedly decrease ALT and AST activity. The enzymatic activity was decreased in a dose-dependent manner in comparison to the group with ethanol alone, which suggests that riboflavin exerts a protective effect against liver injury caused by alcohol. These findings imply the potential therapeutic effect of riboflavin on AH by targeting critical alcohol-metabolizing reactions.
In this study, we identified 64 novel drugs, which are small molecules in nature, with the potential to target AH-involved sub-pathways. We also illustrated the potential mechanisms of the activity of these novel drugs on AH. Although our work needs further experimental verification, the potential molecules identified in this study provide valuable knowledge in the development of novel therapeutic approaches for AH.
Acknowledgments
Funding: This research was supported by the Harbin Medical University Scientific Research Innovation Fund (No. 2016JCZX55), Project of Heilongjiang Health Department (2011-254) and the Undergraduate Student Innovation Project (201810226069).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nagata K, Suzuki H, Sakaguchi S. Common pathogenic mechanism in development progression of liver injury caused by non-alcoholic or alcoholic steatohepatitis. J Toxicol Sci 2007;32:453-68. [Crossref] [PubMed]
- Lucey MR, Mathurin P, Morgan TR. Alcoholic hepatitis. N Engl J Med 2009;360:2758-69. [Crossref] [PubMed]
- Colmenero J, Bataller R, Sancho-Bru P, et al. Hepatic expression of candidate genes in patients with alcoholic hepatitis: correlation with disease severity. Gastroenterology 2007;132:687-97. [Crossref] [PubMed]
- Maddrey WC, Boitnott JK, Bedine MS, et al. Corticosteroid therapy of alcoholic hepatitis. Gastroenterology 1978;75:193-9. [Crossref] [PubMed]
- Dhanda AD, Lee RW, Collins PL, et al. Molecular targets in the treatment of alcoholic hepatitis. World J Gastroenterol 2012;18:5504-13. [Crossref] [PubMed]
- Affò S, Dominguez M, Lozano JJ, et al. Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis. Gut 2013;62:452-60. [Crossref] [PubMed]
- Tilg H, Diehl AM. Cytokines in alcoholic and nonalcoholic steatohepatitis. N Engl J Med 2000;343:1467-76. [Crossref] [PubMed]
- Iimuro Y, Gallucci RM, Luster MI, et al. Antibodies to tumor necrosis factor alfa attenuate hepatic necrosis and inflammation caused by chronic exposure to ethanol in the rat. Hepatology 1997;26:1530-7. [Crossref] [PubMed]
- Mookerjee RP, Sen S, Davies NA, et al. Tumour necrosis factor alpha is an important mediator of portal and systemic haemodynamic derangements in alcoholic hepatitis. Gut 2003;52:1182-7. [Crossref] [PubMed]
- McClain CJ, Barve S, Deaciuc I, et al. Cytokines in alcoholic liver disease. Semin Liver Dis 1999;19:205-19. [Crossref] [PubMed]
- Spahr L, Rubbia-Brandt L, Frossard JL, et al. Combination of steroids with infliximab or placebo in severe alcoholic hepatitis: a randomized controlled pilot study. J Hepatol 2002;37:448-55. [Crossref] [PubMed]
- Tilg H, Jalan R, Kaser A, et al. Anti-tumor necrosis factor-alpha monoclonal antibody therapy in severe alcoholic hepatitis. J Hepatol 2003;38:419-25. [Crossref] [PubMed]
- von Eichborn J, Murgueitio MS, Dunkel M, et al. PROMISCUOUS: a database for network-based drug-repositioning. Nucleic Acids Res 2011;39:D1060-6. [Crossref] [PubMed]
- Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313:1929-35. [Crossref] [PubMed]
- Li C, Li X, Miao Y, et al. SubpathwayMiner: a software package for flexible identification of pathways. Nucleic Acids Res 2009;37:e131. [Crossref] [PubMed]
- Li C, Shang D, Wang Y, et al. Characterizing the network of drugs and their affected metabolic subpathways. PloS one 2012;7:e47326. [Crossref] [PubMed]
- Purohit V, Russo D, Coates PM. Role of fatty liver, dietary fatty acid supplements, and obesity in the progression of alcoholic liver disease: introduction and summary of the symposium. Alcohol 2004;34:3-8. [Crossref] [PubMed]
- Canbay A, Bechmann L, Gerken G. Lipid metabolism in the liver. Zeitschrift fur Gastroenterologie 2007;45:35-41. [Crossref] [PubMed]
- Joeres R, Brachtel D, Gallenkamp H, et al. Caffeine elimination in cirrhotic and non-cirrhotic liver disease of different etiology. Z Gastroenterol 1993;31 Suppl 2:56-61. [PubMed]
- Shiraishi K, Matsuzaki S, Itakura M, et al. Abnormality in membrane fatty acid compositions of cells measured on erythrocyte in alcoholic liver disease. Alcohol Clin Exp Res 1996;20:56A-9A. [Crossref] [PubMed]
- Latour C, Kautz L, Besson-Fournier C, et al. Testosterone perturbs systemic iron balance through activation of epidermal growth factor receptor signaling in the liver and repression of hepcidin. Hepatology 2014;59:683-94. [Crossref] [PubMed]
- Resnick RH, Iber FL. Treatment of acute alcoholic hepatitis. Gut 1972;13:68-73. [Crossref] [PubMed]
- Majumdar SK, Shaw GK, O'Gorman P, et al. Blood vitamin status (B1, B2, B6, folic acid and B12) in patients with alcoholic liver disease. Int J Vitam Nutr Res 1982;52:266-71. [PubMed]
- Vidal F, Lorenzo A, Auguet T, et al. Genetic polymorphisms of ADH2, ADH3, CYP4502E1 Dra-I and Pst-I, and ALDH2 in Spanish men: lack of association with alcoholism and alcoholic liver disease. J Hepatol 2004;41:744-50. [Crossref] [PubMed]


