Identification of key candidate genes and pathways revealing the protective effect of liraglutide on diabetic cardiac muscle by integrated bioinformatics analysis
Introduction
Diabetes mellitus is becoming a major health problem as the International Diabetes Federation (IDF) expects an astounding 642 million diabetes patients by 2040 (1). This chronic metabolic disease causes a series of serious long-term complications affecting the cardiovascular system, peripheral vascular system, central nervous system, and renal function (2). Diabetic cardiomyopathy, among these, exists independently of macro-and micro-coronary artery diseases and other cardiovascular diseases, and presents a high risk of cardiovascular morbidity and mortality (3).
The major pathological manifestations of diabetic cardiomyopathy include hypertrophy, ventricular dilatation, and compromised contractile function, which may be attributed to apoptosis and interstitial fibrosis, leading to ventricular remodeling (1). Previous studies from our lab and others have depicted a few pathophysiological factors that are involved in the onset and development of diabetic cardiomyopathy, including glucose and lipid toxicity, inflammation, oxidative stress, mitochondrial injury, interstitial fibrosis, apoptosis, and dysregulated autophagy (2,3).
Little is known about the physiological process that underlies the progression of diabetic cardiomyopathy, which prohibits adequate treatment. As such, a deeper understanding of the specific processes that govern cell apoptosis and proliferation, as well as the progression of diabetic cardiomyopathy, are crucial for the invention of better therapeutic and diagnostic modalities.
Liraglutide is a glucagon-like peptide-1 (GLP-1) analog that binds to the GLP-1R receptor and has been shown to be abundantly expressed by RGC-5 cells (4). A GLP-1 homolog, exendin [9–39], is 53% similar in sequence homology and is commonly utilized as a GLP-1 receptor antagonist in experimental studies (5). Zhang et al. reported Glp1r expression on retinal ganglion cells and investigated the GLP-1 protective mechanisms against hyperglycaemic conditions (6). The diabetic retina in the early stages of DR is protected by GLP-1 (7). Liraglutide enhances a systemic glycaemic reduction while providing multiple protective effects against diabetic complications (8,9). Despite previous research, the specific role of GLP-1 on diabetic cardiomyopathy has yet to be investigated. Therefore, the current study used the microarray dataset, GSE102194 which was extracted from the NCBI-Gene Expression Omnibus database (NCBI-GEO) (https://www.ncbi.nlm.nih.gov/geo).
The sample size included cardiac muscle samples of 3 liraglutide-treated rats and 3 untreated controls. Differentially expressed genes (DEGs) were identified and used to perform the Gene Ontology and pathway enrichment analyses using the DAVID online tool. Protein-protein interaction (PPI) networks (http://string-db.org) and modular analysis were constructed to discern hub genes. We also found that these modules were associated with cardiac muscle contraction, hypertrophic cardiomyopathy (HCM), and MAPK signaling pathway. We also confirmed that Glp-1 increased H9c2 cell growth after treatment with a high volume of glucose (22 mM, 72 h) using CCK-8 assays. Glp-1 attenuated high glucose-induced cell apoptosis accompanied by a decrease in the level of proinflammatory cytokines (IL-1, IL-1β, and TNF-α) as confirmed by the ELASE kit. Glp-1 also decreased the expression of p38, JNK, ERK1/2, and MARS proteins induced by HG. DEG identification, along with the respective enriched biological pathways and functions, may provide immense insight into the mechanism of liraglutide-protected diabetic cardiomyopathy.
Methods
Microarray data information
GSE102194 gene expression profiles were extracted from the GEO database. GSE102194, which was based on GPL17117 platform Affymetrix Rat Gene 2.0 ST Array) (10). The GSE102194 cohort comprised 6 samples, including the cardiac muscle of 3 liraglutide-treated rats and that of 3 controls.
DEG identification
Raw data files were downloaded in TXT file formats (Affymetrix platform), with subsequent analyses completed on the GEO2R software. Hierarchical clustering analysis was applied to group the data into 2 similar expression patterns of liraglutide-treated and control rat cardiac muscle. Principal component analysis (PCA) in GeneSpring was used to determine probe quality control. The cutoff criterion for statistically significant DEGs was defined as (logFC) >1.
Functional enrichment analysis of DEGs
Candidate DEGs enriched pathways and functions were discerned using the DAVID tool, a freely available online tool that allows gene visualization and annotation as well as integrated discovery function, allowing for a holistic interpretation of the biological function of any one gene (https://david.ncifcrf.gov/) (11). Gene ontology analysis (GO) is a commonly used means of gene and gene product annotation; it is also able to determine the defining biological characteristics of high-throughput genome or transcriptome data (12). The Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.gen ome.jp/) is a database of genetic functions that links both functional and genomic information (13). Both a P<0.01 and FDR<0.05 were taken to indicate statistical significance.
Integration of protein-protein interaction (PPI) network and modular analysis
DEGs were mapped onto the STRING (Search Tool for the Retrieval of Interacting Genes) database (version 9.0) to evaluate inter-DEG relationships via protein-protein interaction (PPI) information (http://string-db.org) (14). Protein interaction relationship networks were mapped using the Cytoscape software (15), which analyzed the relationship between candidate DEG encoding proteins in the cardiac muscle of liraglutide-treated rats. Node degree was calculated using the Network Analyzer plug-in, which involved determining the number of interconnections to sort out PPI hub genes. Molecular Complex Detection (MCODE) was utilized to filter PPI network modules on Cytoscape, selecting only modules with a node number of less than 4 and MCODE scores of more than 4. These nodules were then subjected to pathway and function enrichment analyses. P<0.01 was determined as significantly different.
Cell culture and transfection
H9c2 cells were acquired from the Chinese Academy of Science, Shanghai Cell Bank and cultured in low glucose (5.5 mM) minimum essential medium (Gibco-Invitrogen, Grand Island, NY, USA) complemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco BRL, Grand Island, NY, USA), 100 U/mL of penicillin, and 50 g/mL of streptomycin (Beyotime Institute of Biotechnology, China). The cells were placed in a humidified atmosphere containing 5% CO2 at 37 °C.
To establish a high glucose- (HG-) induced stress model in H9c2 cells, D-glucose (Sinopharm Chemical Reagent Co. Ltd., Shanghai, China) was added in culture medium to reach the final concentration of 22 mM glucose. The concentration of 5.5 mM glucose was used as the control group. A dose-dependent effect of propofol was evaluated by adding 5, 10, 20, and 40 µM of propofol to the cells.
Cell counting Kit (CCK)-8 assay
H9c2 cells were treated with Glp-1 and harvested into 96-well plates for 48 h. CCK8 (Dojindo Molecular Technologies, Japan) was tested at 450 nm by Multiskan Spectrum (Thermo Fisher, USA).
TUNEL assay
TUNEL assay was performed to examine the apoptosis of cells. Specifically, H9c2 cells were fixed with 4% formaldehyde and permeabilized with Cytonin. Streptavidin–fluorescein conjugate was applied to detect the biotinylated nucleotides. Fluorescein-stained cells were analyzed by fluorescence microscopy.
Flow cytometric analysis
The apoptotic assay was conducted using an Annexin V-FITC/PI Apoptosis Detection Kit (KeyGEN Bio-TECH, Jiangsu, China). H9c2 cells were double-stained with Annexin V-FITC and propidium iodide (PI) and analyzed with flow cytometry (FACS- can, BD Biosciences). The relative ratio of early and terminal apoptotic cells was detected to calculate the apoptotic rate.
Enzyme-linked immunosorbent assay (ELISA) of IL-1β, IL-6, and TNF-α
The relative levels of IL-6, IL-1β, and TNF-α in our manuscript were evaluated by the ELISA kits (R&D Systems, USA) as their protocol.
Antibodies and western blot analysis
All proteins were obtained from H9c2 cells, cells were lysed with RIPA buffer, and 12.5% SDS-PAGE was used to separate the proteins. The proteins were transferred to a nitrocellulose membrane (Pall Corporation, USA) and blocked. Next, the blots were incubated with primary antibodies to p38; ERK1/2, JNK, and MRAS were bought from Cell Signaling Technology, USA. After washing and incubating with rabbit or mouse secondary antibodies (LI-COR Biosciences, USA), the blots were with the help of an Infrared Imaging System (LI-COR Biosciences, USA). The densities of bands were quantified using Odyssey 3.0 software. The blots were subjected to densitometry using β-actin as an internal control. The densities of bands from each group were calculated.
Statistical analysis
Data were expressed as means ± SD. Student’s non-paired t-test or one-way analysis of variance (ANOVA) followed by Tukey’s test were used to compare multiple groups. Differences were considered significant when P<0.05. SPSS software (version 13.0; USA) was used to perform the analysis.
Results
DEG identification
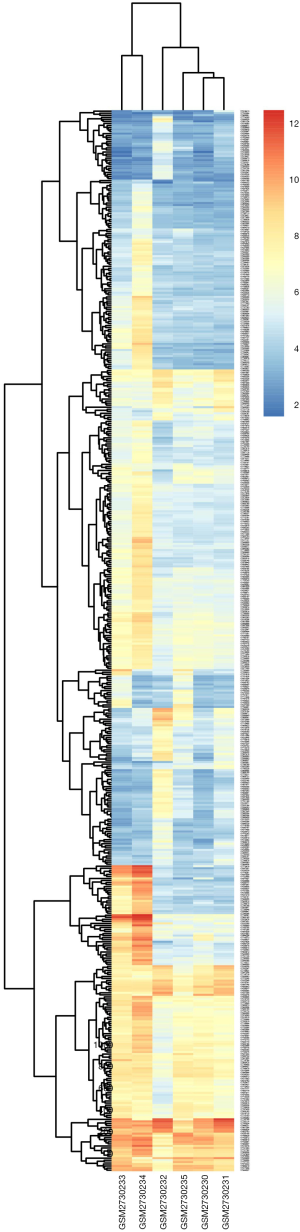
DEGs were identified by separately analyzing microarray data from each chip with the GeneSpring software. Using (logFC) >1 as threshold cutoff point, 520 genes were highlighted. Of these, there were 159 down-regulated genes and 361 up-regulated genes in liraglutide-treated diabetic cardiac muscle in comparison to untreated diabetic cardiac muscle. A heat map of the 520 DEGs using data profile GSE102194 is shown in Figure S1.
GO term enrichment and KEGG pathway analyses
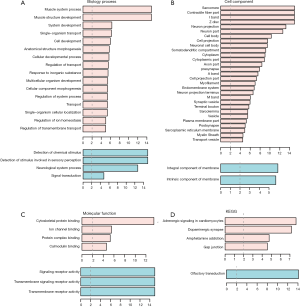
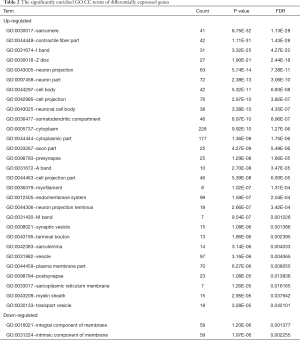
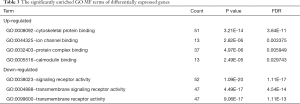
GO and pathway enrichment of DEGs were constructed with the DAVID online databases. The DEGs were categorized based on cellular components, biological processes, and molecular functions. In the biological process group, up-regulated genes were mainly enriched in the muscle system, development of muscle structure, and anatomical structure morphogenesis while the down-regulated DEGs were markedly enriched in biological processes, including the detection of chemical stimulus and neurological system process (Figure 1A and Table 1). GO cell component (CC) analysis also demonstrated significant enrichment of up-regulated DEGs in the sarcomere, contractile fiber, and neuron projection, and significant enrichment of down-regulated DEGs in the integral component of membrane (Figure 1B and Table 2). In addition, for molecular function (MF), the down-regulated DEGs were enriched in signaling receptor activity and transmembrane signaling receptor activity, while the upregulated DEGs were enriched in cytoskeletal protein and ion channel binding (Figure 1C and Table 3).
Full table
Full table
Full table
Table 4 and Figure 1D show the most significantly enriched pathways of the up-regulated and down-regulated DEGs as identified through KEGG analysis. The up-regulated DEGs were enriched in adrenergic signaling in cardiomyocytes, dopaminergic synapse and circadian entrainment, while down-regulated DEGs were enriched in factory transduction.

Full table
Identification of key candidate genes and pathways using protein-protein interaction network (PPI) and modular screening
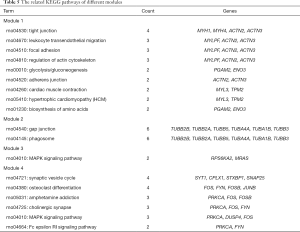
With the STRING online database (Available online: http://string-db.org) and Cytoscape software, 249 DEGs of the 520 DEGs were mapped onto a PPI network, which included 776 edges and 249 nodes (Figure 2A). The top 10 hub nodes with higher degrees were further analyzed. These hub genes included Acta1, Actn2, Mapk12, Tnnc2, Actn3, Eno3, Myh1, Mylpf, Pgam2, and Pygm. Moreover, this network was further assessed with MCODE plug-ins. The top 4 modules from the PPI network were selected for functional annotation. Pathway annotation analysis demonstrated that Module 1 consisted of 17 nodes and 111 edges (Figure 2B, Table 5), which were primarily involved in tight junction, regulation of actin cytoskeleton, glycolysis/gluconeogenesis, and cardiac muscle contraction. Module 2 comprised 7 nodes and 15 edges (Figure 2C, Table 5), which were primarily involved with gap junction and phagosome. Module 3 and Module 4 (Figure 2D,E) were primarily involved in the MAPK signaling pathway and Synaptic vesicle cycle, respectively.

Full table
The effect of GLP-1 regulates the cell viability induced by high-glucose stress
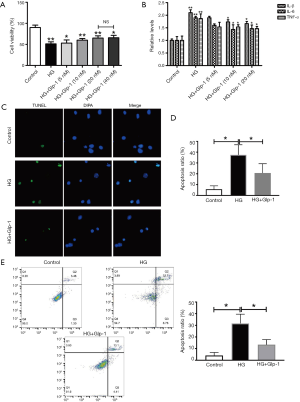
To explore the role of Glp-1 in diabetic cardiac muscle. A diabetic cardiac muscle model was established by H9c2 cells. Treatment with different concentrations of glucose (5.5, 11.0, 22.0, and 44.0 mM) for 72 h. At the concentration of 22 mM, glucose significantly decreased the cell viability of H9c2 (Figure S2). Therefore, 22.0 nM glucose was used as a standard of high glucose-stress for further study. The effect of Glp-1 on the viability of H9c2 cells was measured by CCK8 assay. Compared to the high glucose (22 mM) group, Glp-1 increased H9c2 cell viability in a dose-dependent manner at the range of 5–20 nM. However, 40 nM of Glp-1 did not further improve cell viability during high-glucose insult (Figure 3A).

GLP-1 decreases high-glucose-induced apoptosis in H9c2 Cells.
As shown in Figure 3B, after H9c2 cells were cultured with high glucose (22 mM, 72 h), the levels of IL-6, IL-1β, and TNF-α were significantly increased in the media compared to the control group, whereas, pretreatment of GLP-1 at concentrations of 10 and 20 µM significantly reduced IL-6, IL-1β, and TNF-α expression. Compared to the 10 nM GLP-1 group, the 20 nM Glp-1 group further inhibited inflammatory cytokines. The number of apoptotic cells was labeled with Tunel+. The result showed that the number of apoptotic H9c2 cells cultured with high glucose was significantly increased compared to the control group, whereas pretreatment of GLP-1 at concentrations of 20 µM significantly reduced the glucose-induced apoptotic cells (Figure 3C,D). Similar to our observations, the result of flow cytometric analysis showed the same pattern (Figure 3E). Our data suggest that GLP-1 can protect those H9c2 cells injured by high glucose-induced apoptosis.
GLP1 alleviated apoptosis induced by high-glucose stress in H9c2 Cells by MRAS /MAPK signaling pathway
Our previous results (Table 5) show that MRAS and MAPK signaling pathways might play an essential role in Glp-1-protected H9c2 cells that are injured by high glucose. Compared to the control group, high glucose significantly decreased the protein levels of p38, JNK, EKR1/2, and MRAS. As expected, compared to the model group, the levels of p38, JNK, EKR1/2, and MRAS were increased with high glucose. Furthermore, GLP-1 could inhibit the expression of p38, JNK, EKR1/2, and MRAS induced by high glucose (Figure 4A,B,C,D).

Discussion
Existing literature about the pathophysiological factors of diabetic cardiomyopathy either has only examined a single genetic event or generate results based on a single cohort (16). This lack of information has led towards the stunted progression of knowledge in this field, and indirectly, to the increased mortality and incidence of this deleterious condition. Liraglutide is a novel agent widely used in diabetes mellitus treatment (17). However, the understanding of the relationship between liraglutide and diabetic cardiomyopathy is unclear. Given that microarray analysis and high throughput sequencing provides vast amounts of data on the human genome, these methods have been utilized as a means to predict likely therapeutic targets for liraglutide in diabetic cardiomyopathy treatment.
The current study used the GSE102194 dataset information and successfully extracted 520 DEGs (361 up-regulated and 159 down-regulated) from the cardiac muscle of liraglutide-treated diabetic rats and untreated controls. Functional annotation demonstrated that these DEGs were primarily associated with cardiomyocytes, dopaminergic synapses, and circadian entrainment. Through the construction of PPIs, several important genes were highlighted that may potentially provide insight into the effects of liraglutide on the cardiac muscle of diabetic rats in CRC. Four modules of the PPI were chosen with most of the involved genes found to be enriched in some pathways such as glycolysis/gluconeogenesis, cardiac muscle contraction, MAPK signaling pathway, and synaptic vesicle cycle.
GO BP term analyses revealed that up-regulated DEGs were primarily associated with muscle system processes, the development of muscle structure, and cell development, while down-regulated DEGs were associated with signal transduction and positive regulation of the immune system processes. These findings are consistent with the current theory stating that diabetic cardiomyopathy initiation and progression are highly dependent on the development of muscle structure and muscle system processes, and cell development (18). Additionally, the enriched KEGG pathways of up-regulated DEGs encompassed adrenergic signaling in cardiomyocytes and dopaminergic synapses. Previous studies have shown that adrenergic signaling in cardiomyocyte-related gene upregulation in human cardiomyocyte development wields a predictive ability of the overall survival of cardiomyopathy patients.
PPI networks, including the top degree hub genes of Acta1, Actn2, Mapk12, Tnnc2, Actn3, Eno3, Myh1, Mylpf, Pgam2, and Pygm were also constructed. Acta1 was identified as a hub gene that regulates the non-immunosuppressive immunophilin ligand modifier of gene expression in the dorsal root ganglia during painful diabetic neuropathy (19). Reza et al. (20) reported that ACTA1 was a likely novel pathogenic variant in the dilated cardiomyopathy family. Tadokoro et al. (21) also reported that congenital myopathy demonstrated a fiber-type disproportion in the context of dilated cardiomyopathy in a patient with a novel p.G48A ACTA1 mutation. The second hub gene, Actn2, is a lone muscle isoform of α-actinin that can regulate diabetic cardiomyopathy and is expressed abundantly on cardiac muscle (22). Previous authors have suggested that there is a relationship between diabetic cardiomyopathy and Actn2. Additionally, Actn2 affects the incorporation of cardiomyocyte Z-disc and actin-binding and therefore has a significant role in hypertrophic cardiomyopathy (22). ACTN3 is also a member of the actinin family that acts to cross proteins (23). Moreover, through in vivo co-immunoprecipitation, Eno3 can inhibit fluorescent 3D-co-localization in differentiated H9C2 cells (24). Myh1 has been documented to control muscle fibers, DNA repair, or tumor suppressor (25,26). Of further note, Inal-Gültekin et al. (27) demonstrated Pygm mutations in patients with McArdle disease in a Turkish cohort as determined by next-generation sequencing. Considering the above, additional studies investigating the function of Mylpf in diabetic cardiomyopathy are necessary (28). Nevertheless, the biological functions of Pgam2 in diabetic cardiomyopathy remain unclear.
Modular analysis of the PPI network uncovered that the effect of liraglutide on diabetic rats’ cardiac muscle was associated with cardiac muscle contraction and hypertrophic cardiomyopathy (HCM). Using the STRING online database and Cytoscape software, 249 DEGs of the 520 DEGs were mapped onto a PPI network. We further analyzed the top 10 hub nodes with higher degrees. The PPI network was further assessed using MCODE plug-ins, and the top 4 modules from the PPI network were selected for the functional annotation, which played critical roles in the PPI network. Module 1 was primarily involved in tight junction, regulation of actin cytoskeleton, glycolysis/gluconeogenesis, and cardiac muscle contraction. Module 2 was primarily involved with gap junction and phagosome. Module 3 and Module 4 were primarily involved in the MAPK signaling pathway and synaptic vesicle cycle, respectively. MYL3 and TPM2 are versatile signaling molecules that regulate a diverse range of activities related to muscle growth (29). The third module/cluster consisted of the MAPK signaling pathway. These findings are backed by Higgins et al. who demonstrated MRAS-mediated Noonan syndrome is related to cardiac hypertrophy (30). MRAS also focuses on mineralocorticoid-receptor antagonists (31).
To investigate the role of MRAS/MAPK in glp-1-protected H9c2 cell against high-glucose injury, we first established a high-glucose injury model (32). Our data show (Figure 3) that glp-1 can inhibit H9c2 cell growth induced by high glucose. This finding is similar to that of Guan et al. in whose study Glp-1 attenuated endoplasmic reticulum stress-induced apoptosis in H9c2 cardiomyocytes through GLP-1R/PI3K/Akt pathways (33). Zhang et al. further found that liraglutide inhibits high glucose-induced oxidative stress and apoptosis in neonatal rat cardiomyocytes (34). IL-6 (35), IL-1β (36), and TNF-α (37) had important roles in the progress of the high glucose-induced cardiotoxicity in vitro model. Our data (Figure 3) indicate that glp-1 could decrease the levels of IL-6, IL-1β, and TNF-α that were elevated by high glucose. Interestingly, glp-1 attenuated the high glucose-induced inflammation response by reducing the production of proinflammatory cytokines by inhibiting MRAS/MAPK (Figure 4).
Diabetes mellitus, a chronic metabolic disease, is becoming a significant health threat and triggers a cascade of severe, long-term complications involving the cardiovascular system, peripheral vascular system, central nervous system, and renal function. Ventricular dilatation, hypertrophy, and compromised contractile function are the major pathological manifestations of diabetic cardiomyopathy. Diabetic cardiomyopathy, which occurs independent of macro- and micro-coronary artery diseases and other cardiovascular diseases, imposes a high risk for cardiovascular morbidity and mortality. Our findings elucidated the regulatory network in diabetic cardiomyopathy and might provide a novel diagnostic and therapeutic target for diabetic cardiomyopathy.
To conclude, our study provides a comprehensive bioinformatics analysis of DEGs potentially involved in diabetic cardiomyopathy. The molecular targets uncovered in this study are prime candidates for further investigations into molecular mechanisms and biomarkers.
Acknowledgments
Funding: This work was supported in part by the National Nature Science Foundation of China (No. 81370902, 81170744).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Knudsen JG, Rorsman P. beta Cell Dysfunction in Type 2 Diabetes: Drained of Energy? Cell Metab 2019;29:1-2. [Crossref] [PubMed]
- Dai C, Jiang S, Chu C, et al. Baicalin protects human retinal pigment epithelial cell lines against high glucose-induced cell injury by up-regulation of microRNA-145. Exp Mol Pathol 2019;106:123-30. [Crossref] [PubMed]
- Goedeke L, Perry RJ, Shulman GI. Emerging Pharmacological Targets for the Treatment of Nonalcoholic Fatty Liver Disease, Insulin Resistance, and Type 2 Diabetes. Annu Rev Pharmacol Toxicol 2019;59:65-87. [Crossref] [PubMed]
- Yao Y, Li Q, Wang W, et al. Glucagon-Like Peptide-1 Modulates Cholesterol Homeostasis by Suppressing the miR-19b-Induced Downregulation of ABCA1. Cell Physiol Biochem 2018;50:679-93. [Crossref] [PubMed]
- Chen X, Huang T, Shi Y, et al. A GLP1 receptor agonist attenuates human islet amyloid polypeptideinduced autophagy and apoptosis in MIN6 cells. Mol Med Rep 2019;19:1365-71. [PubMed]
- Zhang Y, Parajuli KR, Fava GE, et al. GLP-1 Receptor in Pancreatic alpha-Cells Regulates Glucagon Secretion in a Glucose-Dependent Bidirectional Manner. Diabetes 2019;68:34-44. [Crossref] [PubMed]
- Liu P, Song J, Liu H, et al. Insulin regulates glucagon-like peptide-1 secretion by pancreatic alpha cells. Endocrine 2018;62:394-403. [Crossref] [PubMed]
- Rakipovski G, Rolin B, Nohr J, et al. The GLP-1 Analogs Liraglutide and Semaglutide Reduce Atherosclerosis in ApoE(-/-) and LDLr(-/-) Mice by a Mechanism That Includes Inflammatory Pathways. JACC Basic Transl Sci 2018;3:844-57. [Crossref] [PubMed]
- Zhu HL, Liu ZP, Yang WY, et al. Liraglutide Ameliorates beta-Amyloid Deposits and Secondary Damage in the Ipsilateral Thalamus and Sensory Deficits After Focal Cerebral Infarction in Rats. Front Neurosci 2018;12:962. [Crossref] [PubMed]
- Zhang Q, Xiao X, Zheng J, et al. Liraglutide protects cardiac function in diabetic rats through the PPARalpha pathway. Biosci Rep 2019. [Epub ahead of print]. [PubMed]
- Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:3. [Crossref] [PubMed]
- Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res 2006;34:D322-6. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2013;41:D808-15. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Zhao X, Huang K, Zheng M, et al. Effect of liraglutide on blood pressure: a meta-analysis of liraglutide randomized controlled trials. BMC Endocr Disord 2019;19:4. [Crossref] [PubMed]
- Nagaike H, Ohara M, Kohata Y, et al. Effect of Dulaglutide Versus Liraglutide on Glucose Variability, Oxidative Stress, and Endothelial Function in Type 2 Diabetes: A Prospective Study. Diabetes Ther 2019;10:215-28. [Crossref] [PubMed]
- Vaikhanskaya TG, Sivitskaya LN, Kurushko TV, et al. Kardiologiia 2018;58:33-45. [Crossref] [PubMed]
- Yamazaki S, Yamaji T, Murai N, et al. FK1706, a novel non-immunosuppressive immunophilin ligand, modifies gene expression in the dorsal root ganglia during painful diabetic neuropathy. Neurol Res 2012;34:469-77. [Crossref] [PubMed]
- Reza N, Garg A, Merrill SL, et al. ACTA1 Novel Likely Pathogenic Variant in a Family With Dilated Cardiomyopathy. Circ Genom Precis Med 2018;11:e002243. [Crossref] [PubMed]
- Tadokoro K, Ohta Y, Sasaki R, et al. Congenital myopathy with fiber-type disproportion accompanied by dilated cardiomyopathy in a patient with a novel p.G48A ACTA1 mutation. J Neurol Sci 2018;393:142-4. [Crossref] [PubMed]
- Haywood NJ, Wolny M, Rogers B, et al. Hypertrophic cardiomyopathy mutations in the calponin-homology domain of ACTN2 affect actin binding and cardiomyocyte Z-disc incorporation. Biochem J 2016;473:2485-93. [Crossref] [PubMed]
- Murphy AC, Young PW. The actinin family of actin cross-linking proteins - a genetic perspective. Cell Biosci 2015;5:49. [Crossref] [PubMed]
- Uys GM, Ramburan A, Loos B, et al. Myomegalin is a novel A-kinase anchoring protein involved in the phosphorylation of cardiac myosin binding protein C. BMC Cell Biol 2011;12:18. [Crossref] [PubMed]
- Ahn JS, Kim DH, Park HB, et al. Ectopic Overexpression of Porcine Myh1 Increased in Slow Muscle Fibers and Enhanced Endurance Exercise in Transgenic Mice. Int J Mol Sci 2018. [Crossref] [PubMed]
- Jin J, Hwang BJ, Chang PW, et al. Interaction of apurinic/apyrimidinic endonuclease 2 (Apn2) with Myh1 DNA glycosylase in fission yeast. DNA Repair (Amst) 2014;15:1-10. [Crossref] [PubMed]
- Inal-Gültekin G, Toptas-Hekimoglu B, Gormez Z, et al. Myophosphorylase (PYGM) mutations determined by next generation sequencing in a cohort from Turkey with McArdle disease. Neuromuscul Disord 2017;27:997-1008. [Crossref] [PubMed]
- Ryan MT, O'Halloran AM, Hamill RM, et al. Polymorphisms in the regulatory region of the porcine MYLPF gene are related to meat quality traits in the Large White breed. Meat Sci 2016;113:104-6. [Crossref] [PubMed]
- Wang Z, Shang P, Li Q, et al. iTRAQ-based proteomic analysis reveals key proteins affecting muscle growth and lipid deposition in pigs. Sci Rep 2017;7:46717. [Crossref] [PubMed]
- Higgins EM, Bos JM, Mason-Suares H, et al. Elucidation of MRAS-mediated Noonan syndrome with cardiac hypertrophy. JCI Insight 2017;2:e91225. [Crossref] [PubMed]
- Yugar-Toledo JC, Modolo R, de Faria AP, et al. Managing resistant hypertension: focus on mineralocorticoid-receptor antagonists. Vasc Health Risk Manag 2017;13:403-11. [Crossref] [PubMed]
- Yang M, Lin Y, Wang Y, et al. High-glucose induces cardiac myocytes apoptosis through Foxo1/GRK2 signaling pathway. Biochem Biophys Res Commun 2019;513:154-8. [Crossref] [PubMed]
- Guan G, Zhang J, Liu S, et al. Glucagon-like peptide-1 attenuates endoplasmic reticulum stress-induced apoptosis in H9c2 cardiomyocytes during hypoxia/reoxygenation through the GLP-1R/PI3K/Akt pathways. Naunyn Schmiedebergs Arch Pharmacol 2019;392:715-22. [Crossref] [PubMed]
- Zhang L, Li C, Zhu Q, et al. Liraglutide, a glucagon-like peptide-1 analog, inhibits high glucose-induced oxidative stress and apoptosis in neonatal rat cardiomyocytes. Exp Ther Med 2019;17:3734-40. [PubMed]
- Ye EA, Steinle JJ. miR-146a suppresses STAT3/VEGF pathways and reduces apoptosis through IL-6 signaling in primary human retinal microvascular endothelial cells in high glucose conditions. Vision Res 2017;139:15-22. [Crossref] [PubMed]
- Hu L, Yang H, Ai M, et al. Inhibition of TLR4 alleviates the inflammation and apoptosis of retinal ganglion cells in high glucose. Graefes Arch Clin Exp Ophthalmol 2017;255:2199-210. [Crossref] [PubMed]
- Sun M, Yang J, Wang J, et al. TNF-alpha is upregulated in T2DM patients with fracture and promotes the apoptosis of osteoblast cells in vitro in the presence of high glucose. Cytokine 2016;80:35-42. [Crossref] [PubMed]