
Expression and prognostic potential of GPX1 in human cancers based on data mining
Introduction
Cancer, a public health challenge, is an important cause of morbidity and mortality around the world (1). Nowadays, cancer incidence and mortality are rapidly growing worldwide, irrespective of the human development level. It has been reported that over 1.5 million individuals were diagnosed with malignant tumors each year, and cancer claimed at least 600,000 people every year in the United States (2). People worldwide are now facing huge problems with the increases in the number of diagnosed, living with, and dying of cancers (3). Notably, lung cancer is the most frequent cancer and the leading cause of cancer death among males, while breast cancer is the most commonly diagnosed cancer and the leading cause of cancer death (4). Despite the rapid improvement of diagnosis and anti-cancer therapies, cancer is still a leading cause of death and results in serious social and economic burdens. Thus, it is urgent to explore the mechanism of carcinogenesis, as well as investigate potential biomarkers of diagnosis, prognosis, and therapy in cancers.
The glutathione peroxidase (GPX) system belongs to a family of phylogenetically related enzymes and takes part in several important biological contexts. GPX can catalyze the reduction of H2O2 and lead to the oxidation of glutathione (GSH), which can be reduced back by GSH reductase via NADPH (5). Glutathione peroxidase-1 (GPX1), an abundantly expressed member of the GPX family, is a widely distributed intracellular protein that presents in all cells (6). GPX1 is closely correlated with tumorigenesis and disease progression. GPX1 expression has been linked to carcinogenic effects in several malignant cancers (7).
Interestingly, researches also reported a positive role in GPX1 that it may prevent carcinogenesis in the initiation phase due to the ability to reduce oxidative DNA mutations (8). Studies also showed that overexpression of GPX1 suppressed the growth of pancreatic cancer cell lines (9). However, high GPX1 activity was associated with high vascular invasion and a lower survival rate in hepatocellular carcinoma (10).
Moreover, high GPX1 expression was also significantly related to poor clinical outcome in oral squamous cell carcinoma (11). The potential mechanism underlying carcinogenesis, mainly due to the ability of GPX1 to prevent oxidant-mediated cytotoxicity in normal cells, may also, in a way, prevent the oxidant-mediated apoptosis in cancer cells (7). Taken above, it is credible that GPX1 is protective or detrimental biological functions according to distinct types of cancer. Therefore, the role of GPX1 in cancers is still unclear to some extent, and further exploration of the correlation between GPX1 expression and survival and prognosis information in cancer patients is needed.
In the present study, we comprehensively analyzed GPX1 expression and association with prognostic values of cancer patients via databases such as the Oncomine, GEPIA, UALCAN, and DriverDBv3. Additionally, to reveal the potential mechanism of GPX1 in cancers, we investigated the functional network of GPX1 using the GeneMANIA and Metascape interactive online web. These findings in our study provided some useful information about the correlation between GPX1 expression and the prognostic values in brain Lower Grade Glioma, Kidney renal papillary cell carcinoma (KIPP), and acute myeloid leukemia (AML), as well as suggested a potential diagnostic biomarker and therapeutic target in certain cancers.
Methods
Oncomine database analysis
The Oncomine database is the largest oncogene chip database and integrated data mining tool that includes 715 gene expression datasets from 86,733 caners and normal samples (12). The mRNA expression differences of the GPX1 gene between tumors and normal tissues in distinct types of cancer were determined within the Oncomine database. The results from the Oncomine are displayed with the Pvalue of 0.01, fold change of 1.5, and gene ranking of all.
GEPIA analysis
Gene Expression Profiling Interactive Analysis (GEPIA) is a commonly used interactive website that plots expression profiles of given genes. GEPIA, which contains 9,736 tumors and 8,587 normal tissues from the TCGA and the GTEx database, performs survival analysis based on gene expression levels according to user-defined sample selections and methods (13). We used GEPIA to find the expression level of the GPX1 gene in distinct types of cancers. The correlation between GPX1 expression and survival, including OS and DFS, in different cancers, was also analyzed by GEPIA. The HR and P or Cox P values from a log-rank test were included in the plot.
UALCAN database analysis
UALCAN is a comprehensive and user-friendly web resource for analyzing cancer data. UALCAN provides access to determine graphs and plots depicting gene expression and survival curves, evaluate promoter DNA methylation information and perform pan-cancer gene expression analysis (14). In this study, we also used UALCAN to find patient survival information in distinct types of cancers based on GPX1 gene expression.
DriverDBv3 database analysis
DriverDBv3 is a cancer omics database that incorporates RNA expression, miRNA expression, methylation, copy number variation, and somatic mutation (15). One of the useful features is that this database allows cancer, gene, and customized analysis, helping researchers analyze the correlations between cancers and driver genes. DriverDBv3 also finds genes and presents them with different molecular features via published bioinformatics algorithms. We used DriverDBv3 to examine the prognostic potential of GPX1 gene expression levels in different human cancers. Survival-relevant with log-rank P value<0.05 was considered significant.
GeneMANIA analysis
GeneMANIA is a commonly used website for constructing protein-protein interaction (PPI) networks and predicting the function of favorite genes (16).This user-friendly online tool can analyze gene or gene lists by the bioinformatics methods, including gene co-expression, physical interaction, gene co-location, gene enrichment analysis, and website prediction. We predicted the function of the GPX1 gene and visualized the gene networks through the GeneMANIA.
Functional enrichment analysis via Metascape and Cytoscape
The interactive genes of GPX1, constructed by GeneMANIA network, were then all input into the Metascape and Cytoscape for further gene annotation and analysis (17). Cytoscape was used to visualize the results of the protein interaction network (18). Metascapewas applied to perform GO and KEGG pathway analysis and MCODE, a plugin in Cytoscape, was used to obtain the core modules of genes associated with GPX1. GO term analysis includes biological processes (BP), cellular component (CC), and molecular function (MF). P value <0.05 was considered as statistically significant.
Results
GPX1 expression levels in distinct types of human cancers
To find differences in GPX1 expression in tumor and normal tissues, the GPX1 gene expression profile across tumor samples and paired normal tissues were determined using the Oncomine database. This database included a total of 456 unique analyses for GPX1. Over-expression of GPX1 was found in cancers based on 38 significant unique analyses and down-expression in 5 analyses. The GPX1 expression was higher in cervical, gastric, kidney cancers, lymphoma, and leukemia compared to the normal samples. In addition, lower expression was seen in lung cancer, lymphoma, and sarcoma (Figure 1A). This result revealed that higher expression of GPX1 was found in most cancers.
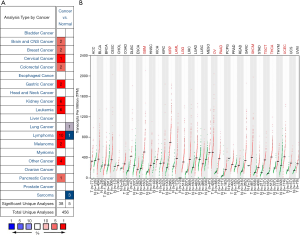
To evaluate the expression differences of GPX1 between tumors and normal tissues in multiple cancers, we further applied the GEPIA website to analyze the data of RNA sequencing expression based on thousands of samples from TCGA and GTEx projects. The result is shown in Figure 1B. The GPX1 expression was higher in GBM (Glioblastomamultiforme), KIRP, LAML (acute myeloid leukemia), LGG (brain lower grade glioma), OV (ovarian serous cystadenocarcinoma), PAAD (pancreatic adenocarcinoma), SKCM (skin cutaneous melanoma), TGCT (testicular Germ Cell Tumors), THCA (thyroid carcinoma), and UCEC (uterine corpus endometrial carcinoma) compared with adjacent normal samples.
Prognostic potential of GPX1 in different cancers
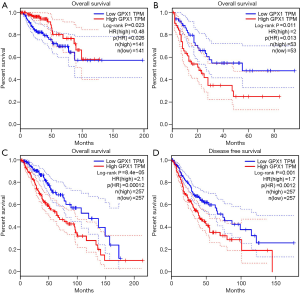
To investigate whether high GPX1 expression level was correlated with prognosis in cancer patients, we used GEPIA to analyze the prognostic value of GPX1 in cancers within the RNA sequencing data in TCGA. Results revealed that high GPX1 expression levels were associated with poorer prognosis of OS and DFS in LGG (Brain Lower Grade Glioma) (OS Log-rank P=8.4e-05, HR =2.1; DFS Log-rank P=0.001, HR =1.7); OS in LAML (Log-rank P=0.011, HR =2). However, high GPX1 expression was correlated with a better prognosis of OS in KIRP (Log-rank P=0.023, HR =0.48) (Figure 2). GPX1 expression has less impact on GBM, OV, PAAD, SKCM, TGCT, THCA, and UCEC. These results revealed that high GPX1 expression leads to a poor prognosis in LGG and LAML patients but a better outcome in KIRP. Meanwhile, the high expression level of the GPX1 gene may have different prognostic value according to distinct types of cancer.
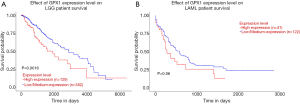
UALCAN database was used to evaluate the prognostic potential of GPX1 in cancer patients further. The relationships between GPX1 expression and significant prognosis information of different cancers are shown in Figure 3. Notably, GPX1 expression significantly affects prognosis in Brain Lower Grade Glioma. However, high expression of GPX1 gene was marginally associated with poorer prognosis in AML (P=0.06). Results showed that high GPX1 expression level was correlated with poor outcome in LGG patients.
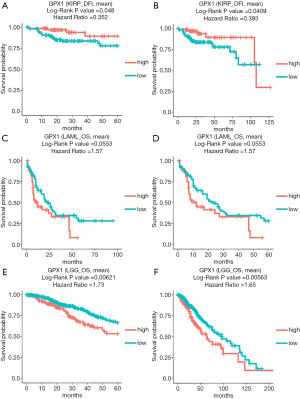
In addition to the analysis of GPX1 in GEPIA and the UALCAN database, the DriverDBv3 database was also used to examine the impact of GPX1 expression on survival rates in different caners. High GPX1 expression was correlated with better prognosis (DFI Log-rank P=0.048, HR =0.352; DFI Log-rank P=0.0409, HR =0.393) in KIRP. The poor prognosis in Brain Lower Grade Glioma (OS Log-rank P=0.00621, HR =1.73; OS Log-rank P=0.00563, HR =1.65) was shown to be correlated with higher GPX1 expression. Meanwhile, high expression of GPX1 in AML was also marginally correlated with poor prognostic value (OS Log-rank P=0.0553, HR =1.57; OS Log-rank P=0.0553, HR =1.57), which was consistent with the result from the UALCAN database. Therefore, these results suggested that GPX1 expression level was a potential prognostic factor in LGG and KIRP patients. However, high GPX1 expression was correlated with a better prognosis in KIRP. Meanwhile, the findings also supplied us with useful information that up-expression of GPX1 also has an impact on the prognosis of AML (Figure 4).
Re-analysis of GPX1 expression in cancers of prognostic values
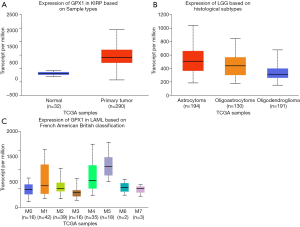
To further examine the expression of GPX1 in cancers showing significant prognostic potential. We performed a subgroup analysis of multiple clinic pathological features of selected cancers in the UALCAN database. Further analysis showed that the transcription level of GPX1 was significantly higher in KIRP patients than normal samples. The expression of GPX1 was significantly higher in LGG patients than healthy people based on histological subgroups analysis. Moreover, high GPX1 expression was found in LAML patients in subgroup analyses based on French American British classification (Figure 5). Thus, it is convincing that GPX1 expression may serve as a potential diagnostic biomarker in these cancer patients.
Promoter methylation levels of GPX1 in cancers of prognostic values
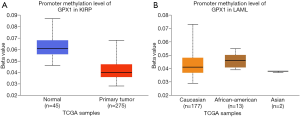
To assess the level of GPX1 promoter methylation in defined types of cancer, we used the UALCAN database to investigate the correlation between GPX1 expression and promoter DNA methylation at the same time. The results suggested that the promoter methylation level of GPX1 in KIRP was lower than normal tissues. Moreover, further sub-group analysis of promoter methylation showed significance based on race in LAML patients. However, GPX1 expression was not significantly associated with promoter DNA methylation in LGG patients (Figure 6). These results revealed that lower promoter DNA methylation might lead to a high expression level of GPX1 in some cancers, such as KIRP.
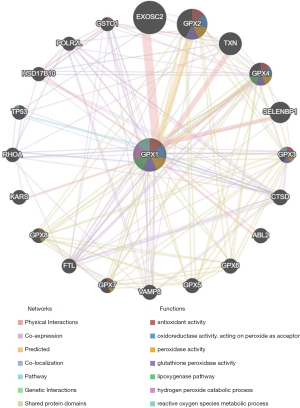
Protein-protein interaction network analysis via GeneMANIA
The protein-protein interaction network revealed a correlation among genes for GPX1. The gene set enriched for GPX1 was responsible for antioxidant activity, oxidoreductase activity, peroxidase activity GPX activity, and reactive oxygen species (ROS) metabolic process (Figure 7).
Functional enrichment analyses of GPX1
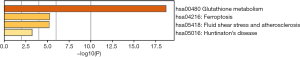
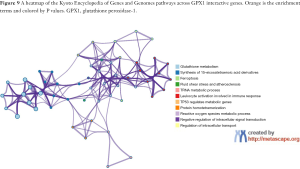
To predict the functional enrichment information of interactive genes of GPX1, GO and KEGG pathway analyses were performed within Metascape. GPX1-associated genes engaged inseveralBP, CCs, and MFs. We found that GPX activity, selenium binding, oxidoreductase activity, tRNA metabolic process, and leukocyte activation involved in immune response had significant regulation by these genes. Moreover, ROSmetabolic process, negative regulation of intracellular signal transduction, and regulation of cell growth also involved in the regulation of GPX1 interactive genes (Figure 8). Significant KEGG pathway analysis showed enrichment in the glutathione metabolism, ferroptosis, fluid shear stress, and atherosclerosis pathways (Figure 9). These findings showed that GPX1 serves an essential role in oxidation metabolism, ferroptosis, ROSmetabolic process, regulation of cell growth, and several other important metabolic processes.

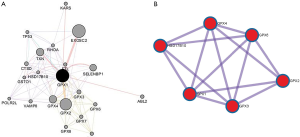
To capture the associations between the enriched terms, we chose a subset of terms and constructed a network plot. Edges linked the terms, including a similarity >0.3. Then, Cytoscape was used to visualize the network. Each node stood for an enriched term colored by the cluster-ID (Figure 10). The enriched terms included the glutathione metabolism, ferroptosis, TP53 regulates metabolic genes, and negative regulation of intracellular signal transduction. Meanwhile, the network of core modules of genes (GPX4, GPX6, GPX2, GPX3), as well as core enriched term (glutathione metabolism) linked to GPX1, were also constructed by MCODE, which indicating important and potential biomarkers that contributed to the development and progression of cancers with GPX1 (Figure 11).
Discussion
Cancers are still threatening human survival around the world despite the great advances in early diagnosis and the emerging correct treatment. KIRP is one of the histological subtypes of epithelial renal cell carcinoma (RCC). Also, KIRP is regarded as a heterogeneous disease in terms of disease progression and clinical outcomes in patients. Glioma is a type of malignancy that originates from the central nervous system and categorizes into four grades (from I to IV) according to the World Health Organization (WHO). Lower-grade gliomas (LGGs) mainly include astrocytoma, oligoastrocytoma, and oligodendroglioma pathological types and represent better prognosis compared with higher grades of gliomas (19). However, LGGs may sometimes unavoidably transform into high grade gliomas despite the benign pathology and comparatively good prognosis. AML is one of the most frequently occurring malignant hematological tumors causing by clonal expansion of undifferentiated myeloid progenitor cells. Most AML patients are adults, and the outcome is still worse compared to younger patients. Relapse is common after chemotherapy due to the minimal residual disease in the bone marrow microenvironment (20). In sum, there is still an urgent need to find potential and useful biomarkers to improve diagnosis, treatment, and prognosis in cancer patients.
The antioxidant enzyme GPX1, commonly distributed in cell mitochondria and cytoplasm, is one of the most notable members of the GPXs family. GPX1 has been reported to play opposite roles in distinct types of cancer. GPX1 overexpression inhibited the growth of pancreatic cancer cells. GPX1 expression was not associated with the radiotherapy response nor survival in head and neck cancer (21). Moreover, the expression level of GPX1 was not associated with prostate cancer recurrence (21,22).
In contrast, high expressed GPX1 tumors were related to poor survival in laryngeal squamous cell carcinoma and esophageal squamous cell carcinoma (23,24). High GPX1 expression promoted the progression and predicted worse overall survival in RCC patients (25).
Moreover, high GPX1 expression has been reported by researchers that could be responsible for chemotherapy resistance in lung cancer, as well as in pancreatic cancer (26,27). Therefore, the expression level of GPX1 could be associated with a different outcome in distinct types of cancer. These findings lead us to extend our study about GPX1, an essential and suitable biomarker for cellular oxidative stress. We investigated the relationships between GPX1 expression and prognostic values in certain human cancers based on data mining.
The present study proved that GPX1 mRNA expression levels were increased in most human cancers. To further investigate the correlation between the GPX1 expression and prognostic values in cancers, we after performing analysis via several online databases. Among them, high expression of GPX1 was found to be significantly associated with the prognostic values in cancers, respectively, KIRP, LGG, and LAML. Recent research revealed that high expression of GPX1 promoted the progression in RCC and GPX1 has the potential to be a biomarker for diagnosis and prognosis of ccRCC patients. In this study, we figured out that the high expression of GPX1 was related to a better outcome in KIRP patients. Also, we re-analyzed the expression of GPX1 in KIRP by UALCAN and found that GPX1 expression in cancer tissues was significantly increased compared with adjacent normal samples. Moreover, the level of GPX1 promoter methylation was decreased in KIRP tissues compared with normal tissues. In sum, these findings suggested that GPX1 may serve as a potential diagnostic and prognostic biomarker in KIRP patients.
Due to the heterogeneity of clinical behavior, LGG cause many morbidities and remain a great therapeutic problem for clinical doctors. Reports showed that the Engrailed 1 (EN1), a member of homeobox-containing transcription factors, can be used as a prognostic marker in LGG patients (28). Besides, the over-expression of selected phosphoprotein 1 (SPP1), an important extracellular glycoprotein, was related to worse overall survival in LGG (29). However, there is little research clarifying the relationship between the GPX1 expression and prognostic potential in LGG. In our study, we found that GPX1 was over-expression in LGG patients compared with normal tissues, and high GPX1 expression was correlated with poor prognosis in LGG.
Meanwhile, the Expression of GPX1 in LGG also showed statistical significance based on histological subtypes, respectively, astrocytoma, oligoastrocytoma, and oligodendroglioma. However, our results showed that GPX1 expression level was not associated with promoter methylation changes in LGG. Therefore, GPX1 over-expression was related to poor survival rate in LGG, and GPX1 may serve as an independent factor in LGG patients.
The leukemogenesis of AML was reported to be associated with increased expression of ROS and GPX1. Also, differentiation agents such as all-trans-retinoic acid (ATRA) could decrease the GPX1 expression in AML primary cell lines, suggesting that GPX1 served as a potential therapeutic target in AML treatment of differentiation agents. This present study proved that GPX1 gene expression levels in AML tissues were higher than that of normal samples. As for promoter methylation changes, we only found out that the promoter methylation level of GPX1 in AML showed significant differences based on race, such as Caucasians and Asians. Meanwhile, we further showed the expression of GPX1 in AML based on French American British classification, and analysis indicated that in some subtypes, the expression level of GPX1 was not significantly different. These results showed that while GPX1 may serve as a potential diagnostic biomarker for AML patients, subtype differences may apply. Therefore, further studies are needed to confirm the role of GPX1 in the diagnosis of AML. Also, this study proved that the overexpression of GPX1 predicted poorer overall survival in patients with AML. However, further analysis in this present study showed a marginal correlation between GPX1 expression and overall survival in AML patients. These results revealed that added clinical samples and wet experiments might be needed to illustrate the prognostic potential of GPX1 in AML. Nonetheless, our results suggested that GPX1 may have an important impact on the survival of patients with AML. The analysis of GPX1 expression, promoter DNA methylation, and survival information prove its useful values as a potential marker of AML patients.
ROS is an important metabolic product in normal cells and has an important impact on most BP such as cell survival, proliferation, and apoptosis. ROS production would increase when exposed to environmental stress. The accumulation of cellular ROS leads to the damage of DNA, thus activating the apoptosis pathway for cell death (30). However, the antioxidant enzyme which defends against oxidative damage can efficiently reduce intracellular ROS. The enzymatic function of GPX1 in the detoxification of lipid and hydrogen peroxides was also regarded as being beneficial in reducing ROS damaging biomolecules such as DNA. In this study, the interaction network of GPX1-associated genes was constructed, and GPX1 was found to be involved in various signaling pathways and BP, including GPX activity, ROS metabolic process, leukocyte activation involved in immune response, regulation of cell growth, and ferroptosis pathways. These findings were consistent with the earlier research.
The biological function of GPX1 was found to be included in the proliferation, invasion, metastasis, apoptosis, and drug resistance in different cancers. The expression of GPX1 was associated with different survival in patients with distinct types of cancer. To our knowledge, GPX4 is also an important member of the GPXs family as GPX1. Researchers have reported GPX4 that it served as an essential regulator of ferroptosis cancer cell death (31). Ferroptosis is a form of nonapoptotic cell death, as well as a mode of cell death involving the production of iron-dependent ROS. Our study suggested that GPX1 may play an essential role in the regulation of cell death and antioxidant via interaction with GPX4. In addition to GPX4, GPX1 was predicted to interact with tumor suppressor TP53 and participated in the TP53 regulation of metabolic genes. These results suggested that GPX1 may be involved in the P53-mediated apoptosis signal pathway via its interaction with TP53. GPX1 was predicted to interact with several essential genes including ABL2, KRAS, and GPX2, which involved apoptosis, immune response, metabolism, and negative signal regulation. These findings showed that GPX1 might take part in a large number of BP in caners by interaction with critical molecules.
Our study used several online databases based on the most popular bioinformatics theories to perform a comprehensive analysis between the genes and tumors. The characteristics of this available method are huge samples, lower cost, and enable large-scale genomics research and functional analyses. However, there are still some limitations to the current study. Firstly, online databases have limitations. Different databases may produce different results due to the various collected sample sizes. The AML tissues and normal control samples have few sample sizes, and a larger number of patients are required to examine the present results. Also, this study only displayed bioinformatics analysis findings based on different online databases. Therefore, further verification experiments including RT-PCR, Western Blot, as well as Immunohistochemical experiments, are needed to verify the findings of the present study.
Conclusions
GPX1 is a member of the GPX family that considered as an enzyme interacting with oxidative stress, whose differential expression is closely related to carcinogenesis and disease progression. In the current study, we conducted a systemic analysis of the expression and the prognostic value of GPX1 in distinct types of cancer. These results suggested that GPX1 overexpression was correlated with significant survival and prognostic values in KIRP, LGG, and LAML patients. Besides, utilizing the comprehensive analysis of various online databases, we finally revealed several enriched pathways like the GPX activity, ferroptosis, regulation of cell growth, and ROS pathways were significantly associated with the GPX1 gene. These findings supplied us multi-level information for the importance of GPX1 in carcinogenesis and potential role as a diagnostic and prognostic biomarker in certain malignant carcinomas. Subsequently, experiments would be performed to further explore the correct molecular mechanisms of GPX1 in cancers of interest.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Pinsky PF. Lung cancer screening with low-dose CT: a world-wide view. Transl Lung Cancer Res 2018;7:234-42. [Crossref] [PubMed]
- Solanki S, Chakinala RC, Haq KF, et al. Inpatient burden of gastric cancer in the United States. Ann Transl Med 2019;7:772. [Crossref] [PubMed]
- White MC, Babcock F, Hayes NS, et al. The history and use of cancer registry data by public health cancer control programs in the United States. Cancer 2017;123 Suppl 24:4969-76. [Crossref] [PubMed]
- Schootman M, Ratnapradipa K, Loux T, et al. Individual- and county-level determinants of high breast cancer incidence rates. Transl Cancer Res 2019;8:S323-33. [Crossref]
- Brigelius-Flohe R, Maiorino M. Glutathione peroxidases. Biochim Biophys Acta 2013;1830:3289-303. [PubMed]
- Lubos E, Loscalzo J, Handy DE. Glutathione peroxidase-1 in health and disease: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal 2011;15:1957-97. [Crossref] [PubMed]
- Brigelius-Flohé R, Kipp A. Glutathione peroxidases in different stages of carcinogenesis. Biochim Biophys Acta 2009;1790:1555-68. [PubMed]
- Baliga MS, Wang H, Zhuo P, Schwartz JL, Diamond AM. Selenium and GPx-1 overexpression protect mammalian cells against UV-induced DNA damage. Biol Trace Elem Res 2007;115:227-42. [Crossref] [PubMed]
- Liu J, Hinkhouse MM, Sun W, et al. Redox regulation of pancreatic cancer cell growth: role of glutathione peroxidase in the suppression of the malignant phenotype. Hum Gene Ther 2004;15:239-50. [Crossref] [PubMed]
- Guerriero E, Capone F, Accardo M, et al. GPX4 and GPX7 over-expression in human hepatocellular carcinoma tissues. Eur J Histochem 2015;59:2540. [Crossref] [PubMed]
- Lee JR, Roh JL, Lee SM, et al. Overexpression of glutathione peroxidase 1 predicts poor prognosis in oral squamous cell carcinoma. J Cancer Res Clin Oncol 2017;143:2257-65. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Liu SH, Shen PC, Chen CY, et al. DriverDBv3: a multi-omics database for cancer driver gene research. Nucleic Acids Res 2020;48:D863-70. [PubMed]
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214-20. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Gittleman H, Sloan AE, Barnholtz-Sloan JS. An independently validated survival nomogram for lower grade glioma. Neuro Oncol 2019. [Epub ahead of print]. [PubMed]
- Zhou S, Liu P, Zhang H. Bioinformatic analysis of the effects and mechanisms of decitabine and cytarabine on acute myeloid leukemia. Mol Med Rep 2017;16:281-7. [Crossref] [PubMed]
- Dequanter D, Dok R, Koolen L, et al. Prognostic Significance of Glutathione Peroxidase Levels (GPx1) in Head and Neck Cancers. Front Oncol 2017;7:84. [Crossref] [PubMed]
- Ekoue DN, Ansong E, Hong LK, et al. GPX1 Localizes to the Nucleus in Prostate Epithelium and its Levels are not Associated with Prostate Cancer Recurrence. Antioxidants (Basel) 2018;7. [Crossref] [PubMed]
- Zhang Q, Xu H, You Y, et al. High Gpx1 expression predicts poor survival in laryngeal squamous cell carcinoma. Auris Nasus Larynx 2018;45:13-9. [Crossref] [PubMed]
- Gan X, Chen B, Shen Z, et al. High GPX1 expression promotes esophageal squamous cell carcinoma invasion, migration, proliferation and cisplatin-resistance but can be reduced by vitamin D. Int J Clin Exp Med 2014;7:2530. [PubMed]
- Cheng Y, Xu T, Li S, et al. GPX1, a biomarker for the diagnosis and prognosis of kidney cancer, promotes the progression of kidney cancer. Aging (Albany NY) 2019;11:12165-76. [Crossref] [PubMed]
- Chen B, Shen Z, Wu D, et al. Glutathione Peroxidase 1 Promotes NSCLC Resistance to Cisplatin via ROS-Induced Activation of PI3K/AKT Pathway. Biomed Res Int 2019;2019:7640547. [PubMed]
- Meng Q, Shi S, Liang C, et al. Abrogation of glutathione peroxidase-1 drives EMT and chemoresistance in pancreatic cancer by activating ROS-mediated Akt/GSK3beta/Snail signaling. Oncogene 2018;37:5843-57. [Crossref] [PubMed]
- Zhu J, Zhang YQ. Engrailed 1 overexpression as a potential prognostic marker in Lower Grade Glioma. PeerJ 2019;7:e7414. [Crossref] [PubMed]
- Chen J, Hou C, Zheng Z, et al. Identification of Secreted Phosphoprotein 1 (SPP1) as a Prognostic Factor in Lower-Grade Gliomas. World Neurosurg 2019;130:e775-85. [Crossref] [PubMed]
- Fan J, Cai H, Tan WS. Role of the plasma membrane ROS-generating NADPH oxidase in CD34+ progenitor cells preservation by hypoxia. J Biotechnol 2007;130:455-62. [Crossref] [PubMed]
- Yang WS. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014;156:317-31. [Crossref] [PubMed]


