
Machine learning for the prediction of severe pneumonia during posttransplant hospitalization in recipients of a deceased-donor kidney transplant
Introduction
The incidence of rejection has decreased and graft survival has increased with the development and administration of immunosuppressants to kidney transplant recipients. However, immunosuppressants simultaneously create risks of infection including nosocomial, opportunistic and community-acquired infection (1-3). As Fishman et al. (2,4) summarized, immunosuppressive therapies contributed to the individual’s risk for infection. Corticosteroids contributed to pneumocystis, bacteria, CMV and BK polyomavirus nephropathy. Mycophenylate mofetil contributed to early bacteria and late CMV infection. Calcineurin inhibitors contributed to viral replication, gingival infections and intracellular pathogens. Lymphocyte depletion induction contributed to herpes virus activation, BK polyomavirus nephropathy, late fungal and viral infections. A comorbid infection, especially opportunistic infection, is very common after solid organ transplantation. It’s reported 25.1% recipients developed opportunistic infection after kidney or simultaneous pancreas-kidney transplant according to a recent study (5). Additionally, infection remains the second cause of death after kidney transplantation during long-term follow-up in a large recent epidemiological study. Among the deceased recipients, 21% died of infection, second to cardiovascular causes (48% of deceased recipients) and pulmonary infection accounts for the majority (45%) of infection-related deaths after transplantation (6). The incidence rate of nosocomial pneumonia was nearly 51/405 (12.6%) (7). Previous studies reported a significantly higher mortality rate in nosocomial pneumonia than in community-acquired pneumonia recipients. The crude mortality rate of nosocomial pneumonia was reported to range from 35–58% and from 3–8% for community pneumonia (8-10). Deceased donors, particularly donors whose organs are accepted based on expanded criteria, might contribute to an increase in the infection risk through intensive immunosuppression or donor-derived nosocomial organisms (3). Among the immunosuppressants, anti-thymocyte globulin, a widely used induction biologic, exerts a lasting T-cell depletion effect and affects the B-cells, NK T-cells and regulatory T cells (11). The T-cell depletion effect usually lasts a few months, with a half-life of approximately 1 month (12). Hence, the transplant recipients are in a net state of immunosuppression. Nosocomial pneumonia caused by bacteria occurs frequently within the first month after surgery due to intensive and destructive suppression of the immune system, which exposes recipients to various pathogens (3). Currently, an effective infection risk classification for is not available for transplant recipients, especially for nosocomial infection. Other predictive models for pneumonia acquired during general abdominal surgery are not applicable for kidney transplant recipients due to the preoperative pulmonary condition of patients with end-stage renal disease and the immunosuppressive status of allograft recipients (13). The identification of recipients at high risk of developing severe pneumonia would effectively enable intensive and targeted prophylactic interventions to be administered. It may reduce the incidence and mortality of severe pneumonia.
Machine learning, a subfield of artificial intelligence, has rapidly developed and probably changes current clinical practice. As summarized by Goldenberg et al. (14), most machine learning algorithms are viewed as mathematical models that map a set of observed variables (i.e., features or predictors) into a set of outcome variables (i.e., labels or targets). Machine learning is classified into three paradigms based on the targets: supervised, unsupervised and reinforcement learning. Labels are included in the training dataset in supervised learning, but not in unsupervised learning. Reinforcement learning does not require any data to be provided in advance, but obtains learning information and updates model parameters by receiving rewards or feedback from the environment. Supervised learning algorithms are trained to decrease the predictive error between predictive targets and the ground truth. These algorithms have been subdivided into two categories based on label characteristics: classification (discrete classes) and regression (continuous value label). The prevailing algorithms include linear and logistic regression analyses, a Naïve Bayes classifier, support vector machine (SVM), AdaBoost, Random Forest, Decision tree, etc. The best algorithm for all cases has not been established. The best model usually depends on the sample data set and the purpose of the analysis in a particular scenario. Machine learning algorithms display improved predictive performance and low error in certain scientific fields (15,16).
In this study, we chose the algorithms with classical methods and excellent application practices to design an analytical method. Support Vector Machine is an excellent technology with independent integrity theory based on a global optimal solution (17). Random Forest is a mature technology based on the extensive application of integrated algorithms (18). Logistic regression is a practical technology with a long history based on statistics. Naïve Bayes is an analytical method based on conditional probability, which is effective in predicting most datasets (19). AdaBoost is an enhanced learning algorithm designed to transform weak learners into strong learners with a high prediction accuracy (20). Therefore, in order to identify severe pneumonia recipients, we aim to use these five algorithms to predict severe pneumonia episodes in recipients of deceased donor kidney transplants during the perioperative period and to compare their performance by analyzing our monocentric dataset from 2015 to 2018.
Methods
Study population
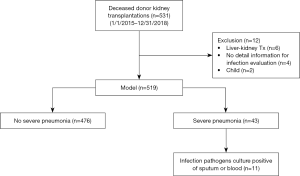
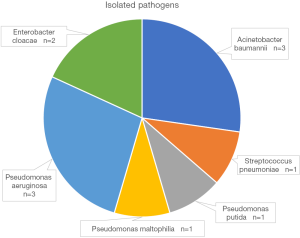
This study was approved by the research ethics committee at the Third Affiliated Hospital of Sun Yat-sen University {IRB approval: [2019]02-512-01}. Allograft kidney transplantation records were retrospectively reviewed from the electronic medical records system of the Third Affiliated Hospital of Sun Yat-sen University and our institute’s access to the China Organ Transplant Response System (COTRS) between January 2015 and December 2018. Five hundred thirty-one transplantation records were retrieved. Patients who met any of the following criteria were excluded: patients who received simultaneous liver and kidney transplantation (n=6), pediatric recipients (n=2), a lack of detailed medical examination records for the infection evaluation (missing values, n=4). Figure 1 displays the screening process. We eventually included 519 transplantations from deceased donors. Of these transplant recipients, forty-three patients (8.3%) suffered severe pneumonia during the perioperative period after transplantation. Among the patients with severe pneumonia, positive microbial isolation was reported in eleven recipients. The major pathogens that were isolated are listed in Figure 2. Nine (20.9%) recipients died of severe pneumonia or related complications. We reported the data in this study in a manner compliant with the RECORD statement (an extension of STROBE reporting guidelines for a study based on routinely-collected health data) (21). All donor sources were deceased citizen donors, since it has been the sole legal method for the procurement of solid organ transplant from a non-relative in mainland China since January 1, 2015. All procured organs were numbered, matched and allocated at the China Organ Transplant Response System (https://www.cot.org.cn/), which is the sole legitimate official site designated by the National Health Commission of China.
Data collection and definition of variables
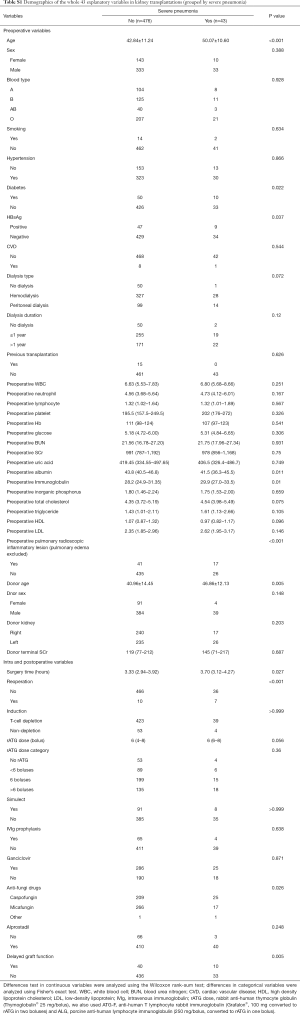
In this study, basic demographics included age, sex, blood type, HBsAg status, smoking status, diabetes, preoperative dialysis, dialysis periods, donor type, previous transplantation history, operation duration, induction therapy, rATG dose, IVIg prophylaxis, ganciclovir prophylaxis, preoperative white blood cell count, neutrophil count, lymphocyte count, serum albumin level, immunoglobulin level, preoperative chest radioscopic screen, delayed graft function, etc. Detailed information for the 43 variables are listed in Table S1. Regarding the variable of reoperation, reoperation after pneumonia occurrence was labeled as no reoperation in this study. Data were obtained from the electronic medical records system, laboratory report system and Picture Archiving and Communication Systems (PACS). The inpatient number was the unique identifier. None of the included patients were infected with HIV infection. Almost all screened recipients were CMV IgG antibody positive after an examination at our local clinical practice. Thus, the serum CMV status was not evaluated in this study.
Full table
Pneumonia was diagnosed according to local hospital-acquired pneumonia guidelines modified based on IDSA/ATS 2016 guidelines, in which pneumonia was defined as a new lung infiltrate and clinical evidence that the infiltrate is of an infectious origin, which includes the new onset of fever, purulent sputum, leukocytosis, and a decrease in oxygenation (22). Due to the lack of accepted criteria for a severe nosocomial pneumonia diagnosis, severe pneumonia was defined according to the IDSA/ATS 2007 criteria for severe community-acquired pneumonia (23). In this study, the minor criterion of uremia in the IDSA/ATS 2007 guidelines was discarded because patients were diagnosed with ESRD. Induction therapy was categorized as the presence or absence of lymphocyte depletion. Delayed graft function was defined as a requirement for posttransplant dialysis (at least one dialysis session during the first 7 days after transplantation) after excluding dialysis for hyperkalemia or fluid overload, etc. (24).
Feature selection
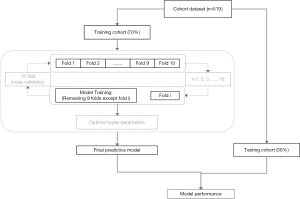
Recursive feature elimination (RFE) selected features by recursively considering increasingly smaller feature sets. First, the classifier was trained on the set of initial features, and the importance of each feature was obtained based on feature importance attributes. Then, the least important feature was deleted from the current set of features. This process was repeated recursively on the pruning set until the required number of functions was finally achieved. We choose the RFE cross-validation algorithm (25) (see Python's Scikit-Learn module), which performed the RFE algorithm in the cross-validation cycle, to identify the optimal number of important features related to the target variables. The selection process is shown in Figure 3.
Model construction
In this study, we chose a tree-based ensemble classification algorithm (Random Forest) to build models based on the different combinations of variables described above. We also used SVM and a Logistic Regression Classifier to build a prediction model based on the combination of the aforementioned variables. The performance of ensemble models and non-ensemble models was compared.
We divided the original data into a 70% training set and 30% test set to obtain a reliable evaluation and avoid overfitting (26). In the training set, the best model parameters were selected through 10-fold cross validation. Then, the model was retrained using the training cohort, and the final prediction model was obtained. Finally, using the final training model, six independent validations were performed on the original data. The detailed process of model selection is shown in Figure 3.
Based on the ideas described above, we began to train machine learning algorithms (Random Forest Classifier, SVM and Logic Regression Classifier). First, preoperative pulmonary lesions (preoperative pulmonary radioscopic inflammatory lesions), preoperative immunoglobulin level, reoperation, recipient age, delayed graft function, preoperative albumin level, donor age and rATG dose were selected to build the SVM classifier model, Logistic Regression classifier model, AdaBoost model, Naïve Bayes model and Random Forest classifier, respectively. The machine learning algorithm was implemented using Python 3.7.4 (https://www.python.org) with Scikit-learn (https://scikit-learn.org/stable/). The predictive performance was compared by calculating the area under the receiver operating characteristic curve (AUROC) and area under the precision-recall curve (AUPRC) for the test dataset (30% of the original cohort, randomly selected samples). Considering the imbalance of labels (targets), we mainly used the AUPRC to evaluate the predictive performance (27).
Statistical analysis
Continuous variables are presented as the means and standard deviations or medians and interquartile ranges, and categorical variables are presented as frequencies and percentages. Differences between groups were analyzed using Fisher’s exact probability test for categorical variables and Welch’s t-test (or the Wilcoxon rank-sum test) for continuous variables. Statistical analyses were performed using Stata/IC version 16.0 (College Station, TX, StataCorp LLC) and R version 3.6.1 (R Foundation for Statistical Computing). A P value <0.05 was considered statistically significant.
Results
Population demographics
Five hundred nineteen kidney transplantation records were included. Among the 519 transplantations, we identified forty-three severe pneumonia episodes, according to the definition. We included 43 explanatory variables in this study. The distribution of patients subdivided by severe pneumonia is presented in Table 1. The whole table is presented in Table S1. Significant differences in recipient age, diabetes status, HBsAg level, preoperative albumin and immunoglobulin levels, preoperative pulmonary lesions, surgery time, reoperation, anti-fungal drug usage and delayed graft function, as well as donor age, were observed between patients with and without severe pneumonia (P<0.05). A further analysis of recipients who died and survived after acquiring severe pneumonia did not reveal significant differences in the aforementioned features, except for the preoperative serum phosphate level. Recipients whose cause of death was severe pneumonia had a higher preoperative phosphate level (P=0.044).
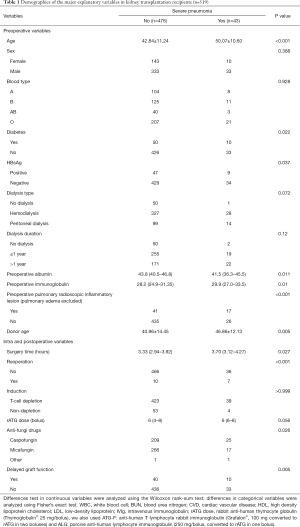
Full table
Feature importance and selection
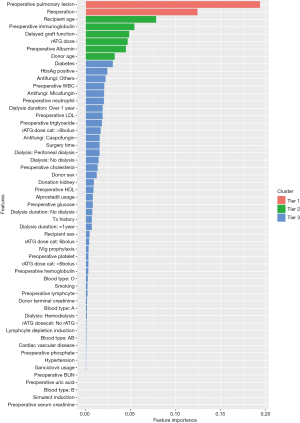
According to the RFECV algorithm of 10-fold cross validation mentioned in the methods section, the feature scores of all variables were calculated in the training cohort (Figure 4 shows the feature importance). Among these features, the top two variables (preoperative pulmonary lesions and reoperation, with importance scores of 0.194 and 0.124, respectively) were the most important. The next echelon of variables included the recipient age, preoperative immunoglobulin level, delayed graft function, rATG dose, preoperative albumin level, and donor age. All of these features displayed a strong correlation with severe pneumonia.
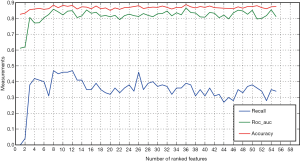
In Figure 5, the sensitivity (recall) improved when the Random Forest model was used in the training cohort, along with the increase of ranked features. When eight variables were included, the best AUROC and sensitivity were achieved. Meanwhile, the predictive accuracy slightly increased from the high baseline performance. The high baseline performance was potentially attributed to the highly imbalanced datasets. The rapid increase in the sensitivity and AUROC with the inclusion of several frontier variables implied the importance of the top-ranked features. In this study, the important features were preoperative pulmonary lesions, reoperation and recipient age.
Model evaluation and comparison
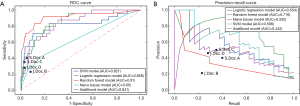
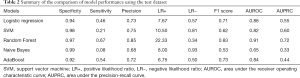
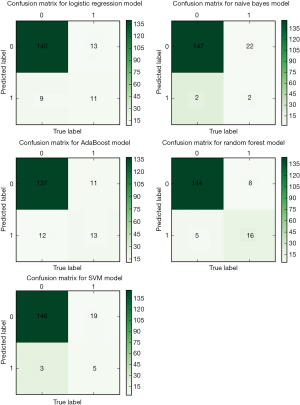
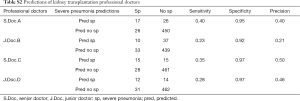
For the selection of a better predictive model, we compared several widely applied machine learning algorithms including support vector machine, Random Forest, Naïve Bayes and AdaBoost, using a receiver operating characteristic curve and precision-recall curve (Figure 6). Both curves revealed the best predictive performance for the Random Forest model, with an AUROC of 0.91 and AUPRC of 0.72. Table 2 summarizes the model metrics, including specificity, sensitivity, precision (positive predictive value, PPV), positive likelihood ratio (LR+, higher is better), negative likelihood ratio (LR−, lower is better), F1 score (higher is better), AUROC, and AUPRC (higher is better). The Random Forest model exhibited the highest sensitivity (0.67), PPV (0.85), LR+ (22.33), AUROC (0.91), AUPRC (0.72) and lowest LR− (0.34). The Naïve Bayes model exhibited the best specificity (0.99). The Random Forest model also displayed a good specificity (0.97). Figure 7 presents the confusion matrix of each model used to predict the test set. The test sample used for model evaluation was oversampled from the original test dataset (30% of the cohort), with 24 targets and 149 negative labels. The predictions of the five machine learning algorithms are shown. We also blindly compared the predictive ability of professional doctors in our kidney transplantation center (Table S2). Two junior doctors presented sensitivity and specificity values of 0.23 and 0.92, and 0.28 and 0.97, respectively. Two senior doctors presented values of 0.40 and 0.95, and 0.35 and 0.97, respectively (shown in Figure 6).
Full table
Full table
Discussion
Infection, particularly pneumonia, is a crucial complication occurring after kidney transplantation. Due to the requirement for immune suppression, particularly using immune induction therapy which made the immune system impotence in the first month, mild pneumonia may progress to severe pneumonia and even lethal sepsis. In our transplantation center, the regular prophylaxis used to combat infection was universally performed to prevent bacterial, fungal and viral amplification. For the prevention of bacterial infections, we regularly prescribed cefoperazone or other sensitive antibiotics according to the donor condition, local bacterial spectrum (gram-negative bacteria, GNB, are pathogens that are frequently isolated from patients with pneumonia) and perfusate culture results. For the prevention of fungal infections, we regularly used echinocandin (caspofungin or micafungin) or other triazole antifungal drugs, according to the culture results. Trimethoprim-sulfamethoxazole (TMP/SMZ) was prescribed to prevent Pneumocystis carinii pneumonia after discharge. Universal cytomegalovirus (CMV) prophylaxis treatment starts after surgery or discharge, and ganciclovir or valganciclovir are applied to prevent CMV replication. Forty-three (8.3%) severe pneumonia episodes still occurred, even after this strong prophylaxis. Nine of the 43 (20.9%) patients died of sepsis or related complications. This result was consistent with previously reported data (10). These findings indicate the importance and necessity of identifying transplant recipients at high risk of developing severe pneumonia. We also analyzed the features of those deceased recipients. A significantly higher preoperative serum phosphate level was observed in recipients who died of severe pneumonia. The phosphate level reflected the adequacy of dialysis to some extent (28). Inadequate dialysis correlates with decreased lung function (29) and potentially contributes to increase the mortality risk (30). In our clinical practice, of course, prophylaxis is one of the most effectively protective modalities. When recipients were diagnosed with severe pneumonia, we will adopt several steps. First, preemptively upgrading antibiotic therapy is emerged. Including broad-spectrum antibiotic like carbapenems and antifungal therapy. Whether supplied antivirus therapy including against CMV or influenza was dependent on the signs of viral pneumonia including clinical manifestations and interstitial pneumonia lesion in radioscopy, as well as for pneumocystis jirovecii. Second, oral immunosuppressants were ceased and instead by methylprednisolone 40 mg bid. Yang et al. (31) summarized their cases and showed no influence on graft function and graft survival after transient cease using of oral immunosuppressants for pneumonia treatment. Third, infection pathogens isolation and multidrug-resistant (MDR) identification was critical important. This was compliant with the diagnosis and treatment guidelines of nosocomial pneumonia. It would effectively guide the usage of anti-infection therapy. Fourth, anti-infection supplemental therapy, like IVIg usage, may improve recovery. But there is no definite evidence to support it. Fifth, monitoring including blood gas, procalcitonin, Pneumonia Severity Index (PSI), and other correlated test could help to guide the treatment. And the last, also the most helpful, was multi-disciplinary team (MDT) including respiratory department, infection department and ICU for pathogen diagnosis and treatment.
In this monocentric study, we reported the predictive performance of machine learning algorithms in identifying severe pneumonia after a deceased donor kidney transplantation. We screened 519 transplantations and identified 43 episodes of severe pneumonia. From the 43 explanatory variables, we selected the eight top-ranked variables through RFECV method for model building. The Random Forest classifier displayed better predictive performance than support vector machine, Naïve Bayes, AdaBoost and the classical statistical-based logistic regression algorithms. It showed an AUROC of 0.91, a sensitivity of 0.67, a PPV of 0.85 and an AUPRC of 0.72 using the top eight features. Meanwhile, the traditional logistic regression-based algorithm also showed impressive performance compared with other machine learning algorithms, including SVM, AdaBoost, Random Forest, and Naïve Bayes. The results of this study once again verify its good application.
Based on the Random Forest model, the top two important features were preoperative pulmonary lesions and reoperation. The next echelon of variables included recipient age, preoperative immunoglobulin level, rATG dose, delayed graft function and preoperative albumin level, the importance of these features was clinically explainable. Obviously, the top three features were correlated with pneumonia episodes. A potential preoperative pulmonary inflammatory lesion tends to indicate a potential asymptomatic infection. Immunosuppression might rapidly deteriorate the infection status. Reoperation was shown to be risk factor for graft survival and early infection (32-34). Age is a significant risk factor for postoperative pneumonia in patients undergoing noncardiac surgery (13). The albumin level is significantly associated with death from pneumonia in recipients of kidney transplants (9,35). In another study, both baseline and posttransplant hypogammaglobulinemia were independent risk factors for infection (36). Our results revealed the importance of the preoperative immunoglobulin level for predicting severe pneumonia. On the other hand, in our dataset, a constant rATG dose is not used in clinical practice. We attempted to follow the recommendation of a total dose of 6 mg/kg divided into three days of administration or even 1.5 mg/kg/d for 3–5 days (37), but massive nosocomial pneumonia emerged. Thus, we reduced the induction dosage according to each recipient’s weight and clinical status after balancing the rejection and infection risk (38). Hence, the use of the rATG dose as a pneumonia predictor was reasonable. Regarding delayed graft function, previous studies had indicated a correlation between graft function and infection risk (34,39). According to the study by Menezes, DGF represents an independent risk factor for surgical site infection (34). In clinical practice, patients with DGF are susceptible to developing various complications, including pulmonary infection. All of the top predictive features described above have clinically reasonable explanations.
In recent years, machine learning has rapidly developed along with advances in computer science. Machine learning-based data mining coexists with traditional statistical methods in medical research. Machine learning focuses on prediction, while traditional statistical methods are mainly used to analyze the causal inference by combining clinical characters and designations. The development of machine learning algorithms contributes to its increasing application in industry and other scientific fields. Compared with traditional statistical predictive models, machine learning algorithms show better predictive performance, particularly for highly dimensional massive data, which traditional statistical models are unable to analyze. In the present study, we used several prevailing algorithms to identify severe pneumonia episodes after kidney transplantation during perioperative period. Our raw dataset showed an obvious imbalance with 43 target events (severe pneumonia) and 476 negative samples. This type of imbalance is very common in clinical research. Thus, we adopted ten-fold cross-validation and an oversampling method to improve the model stability and generalization and to reduce the effect of the imbalance.
For model selection, the AUROC is a general metric. However, AUROC is not a unique reference. In clinical practice, model selection depends on the clinical objective. For example, when a certain target should be completely identified due to its particularity, such as serious lethality, a model with high sensitivity (recall) may be more appropriate. When a relatively high sensitivity is required, a ROC with lower false positive rate (1-specificity) or higher positive predictive value (PPV) may be more appropriate. Thus, the ROC shape is also a reference. For machine learning, the AUPRC and F1 score are practical metrics, particularly for an imbalanced dataset (27). In the present study, we selected a model with a high sensitivity and PPV due to the importance of severe pneumonia. AUPRC would be a good metric and the ROC shape could be referenced. Based on this requirement, we preferred the Random Forest model rather than a logistic regression model. The Random Forest model was deemed an evergreen in machine learning algorithms. Numerous similar medical studies have indicated the superiority and stability of the Random Forest model (27,40).
This study has some limitations. First, all of the aforementioned models were based on a retrospective cohort that may have some level of bias. A prospective external validation cohort is needed for further confirmation. Second, our study was devoted to the predictive performance rather than a statistical inference. Thus, conclusions of conclude causality or risk factors were unable to be determined using machine learning models. Third, our raw dataset had a limitation of a small sample size and target imbalance. We used the oversampling method to generate new synthetic samples from the severe pneumonia recipients, which might affect the validity of the model. Finally, some features would potentially improve the prediction performance, such as HLA mismatch, warm and cold ischemia time and obesity, which require further exploration. Predictive models with large number of explanatory features are not very concise for clinical applications, but might be useful in a highly informationized medical facility.
Conclusions
Machine learning algorithms exhibit an improved predictive performance for severe pneumonia episodes during perioperative periods after kidney transplantation. Among these algorithms, the Random Forest model shows the highest sensitivity, AUROC and AUPRC. Further prospective multicenter studies with multiple datasets are needed to confirm our results and to reduce the influence of the imbalance in the target variables.
Acknowledgments
We acknowledge colleagues from Fane Data Technology Corporation for providing technical support in machine learning model building.
Funding: This study was supported by the National Natural Science Foundation of China (81470977 and 81970652), Ten-Five Project of the Third Affiliated Hospital of Sun Yat-sen University (SW201904), The Third Affiliated Hospital of Sun Yat-Sen University Clinical Research Program (YHJH201906), and Guangzhou Science and Technology Plan Projects (201803010016).
Footnote
Conflicts of Interest: The authors declare no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the research ethics committee at the Third Affiliated Hospital of Sun Yat-sen University {IRB approval: [2019]02-512-01}. All organs were obtained from donations by deceased citizens. The research is compliant with the Declaration of Helsinki and Declaration of Istanbul.
References
- O'Shea DT, Humar A. Life-threatening infection in transplant recipients. Crit Care Clin 2013;29:953-73. [Crossref] [PubMed]
- Fishman JA. Opportunistic infections--coming to the limits of immunosuppression? Cold Spring Harb Perspect Med 2013;3:a015669. [Crossref] [PubMed]
- Fishman JA. Infection in solid-organ transplant recipients. N Engl J Med 2007;357:2601-14. [Crossref] [PubMed]
- Fishman JA, Issa NC. Infection in organ transplantation: risk factors and evolving patterns of infection. Infect Dis Clin North Am 2010;24:273-83. [Crossref] [PubMed]
- Helfrich M, Dorschner P, Thomas K, et al. A retrospective study to describe the epidemiology and outcomes of opportunistic infections after abdominal organ transplantation. Transpl Infect Dis 2017;19:e12691. [Crossref] [PubMed]
- Kinnunen S, Karhapaa P, Juutilainen A, et al. Secular Trends in Infection-Related Mortality after Kidney Transplantation. Clin J Am Soc Nephrol 2018;13:755-62. [Crossref] [PubMed]
- Yang Y, Ren L, Zhang Y, et al. Changes in biochemical parameters on the first day after kidney transplantation: risk factors for nosocomial infection? Chin Med J (Engl) 2010;123:563-8. [PubMed]
- Cervera C, Agusti C, Angeles Marcos M, et al. Microbiologic features and outcome of pneumonia in transplanted patients. Diagn Microbiol Infect Dis 2006;55:47-54. [Crossref] [PubMed]
- Dizdar OS, Ersoy A, Akalin H. Pneumonia after kidney transplant: incidence, risk factors, and mortality. Exp Clin Transplant 2014;12:205-11. [PubMed]
- Hoyo I, Linares L, Cervera C, et al. Epidemiology of pneumonia in kidney transplantation. Transplant Proc 2010;42:2938-40. [Crossref] [PubMed]
- Mohty M. Mechanisms of action of antithymocyte globulin: T-cell depletion and beyond. Leukemia 2007;21:1387-94. [Crossref] [PubMed]
- Bunn D, Lea CK, Bevan DJ, et al. The pharmacokinetics of anti-thymocyte globulin (ATG) following intravenous infusion in man. Clin Nephrol 1996;45:29-32. [PubMed]
- Arozullah AM, Khuri SF, Henderson WG, et al. Development and validation of a multifactorial risk index for predicting postoperative pneumonia after major noncardiac surgery. Ann Intern Med 2001;135:847-57. [Crossref] [PubMed]
- Goldenberg SL, Nir G, Salcudean SE. A new era: artificial intelligence and machine learning in prostate cancer. Nat Rev Urol 2019;16:391-403. [Crossref] [PubMed]
- Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 2017;318:2199-210. [Crossref] [PubMed]
- Meyer A, Zverinski D, Pfahringer B, et al. Machine learning for real-time prediction of complications in critical care: a retrospective study. Lancet Respir Med 2018;6:905-14. [Crossref] [PubMed]
- Huang S, Cai N, Pacheco PP, et al. Applications of Support Vector Machine (SVM) Learning in Cancer Genomics. Cancer Genomics Proteomics 2018;15:41-51. [PubMed]
- Marchese Robinson RL, Palczewska A, Palczewski J, et al. Comparison of the Predictive Performance and Interpretability of Random Forest and Linear Models on Benchmark Data Sets. J Chem Inf Model 2017;57:1773-92. [Crossref] [PubMed]
- Zhang Z. Naive Bayes classification in R. Ann Transl Med 2016;4:241. [Crossref] [PubMed]
- Zhang PB, Yang ZX. A Novel AdaBoost Framework With Robust Threshold and Structural Optimization. IEEE Trans Cybern 2018;48:64-76. [Crossref] [PubMed]
- Benchimol EI, Smeeth L, Guttmann A, et al. The REporting of studies Conducted using Observational Routinely-collected health Data (RECORD) statement. PLoS Med 2015;12:e1001885. [Crossref] [PubMed]
- Kalil AC, Metersky ML, Klompas M, et al. Management of Adults With Hospital-acquired and Ventilator-associated Pneumonia: 2016 Clinical Practice Guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis 2016;63:e61-e111. [Crossref] [PubMed]
- Mandell LA, Wunderink RG, Anzueto A, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis 2007;44 Suppl 2:S27-72. [Crossref] [PubMed]
- Schröppel B, Legendre C. Delayed kidney graft function: from mechanism to translation. Kidney Int 2014;86:251-8. [Crossref] [PubMed]
- Darst BF, Malecki KC, Engelman CD. Using recursive feature elimination in random forest to account for correlated variables in high dimensional data. BMC Genet 2018;19:65. [Crossref] [PubMed]
- Luo W, Phung D, Tran T, et al. Guidelines for Developing and Reporting Machine Learning Predictive Models in Biomedical Research: A Multidisciplinary View. J Med Internet Res 2016;18:e323. [Crossref] [PubMed]
- Chiew CJ, Liu N, Wong TH, et al. Utilizing Machine Learning Methods for Preoperative Prediction of Postsurgical Mortality and Intensive Care Unit Admission. Ann Surg 2019. [Epub ahead of print]. [Crossref]
- Perl J, Dember LM, Bargman JM, et al. The Use of a Multidimensional Measure of Dialysis Adequacy-Moving beyond Small Solute Kinetics. Clin J Am Soc Nephrol 2017;12:839-47. [Crossref] [PubMed]
- Zhang P, Wu HM, Shen QY, et al. Associations of pulmonary function with serum biomarkers and dialysis adequacy in patients undergoing peritoneal dialysis. Clin Exp Nephrol 2016;20:951-9. [Crossref] [PubMed]
- Szeto CC, Wong TY, Chow KM, et al. Impact of dialysis adequacy on the mortality and morbidity of anuric Chinese patients receiving continuous ambulatory peritoneal dialysis. J Am Soc Nephrol 2001;12:355-60. [PubMed]
- Yang B, Ding X, Xie J, et al. Transient stopping immunosuppressive agents during the post-transplant pulmonary infection does not affect the long-term outcome of renal transplantation. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2015;40:380-6. [PubMed]
- Freire MP, Antonopoulos IM, Piovesan AC, et al. Amikacin prophylaxis and risk factors for surgical site infection after kidney transplantation. Transplantation 2015;99:521-7. [Crossref] [PubMed]
- Soong RS, Chan KM, Chou HS, et al. The risk factors for early infection in adult living donor liver transplantation recipients. Transplant Proc 2012;44:784-6. [Crossref] [PubMed]
- Menezes FG, Wey SB, Peres CA, et al. Risk factors for surgical site infection in kidney transplant recipients. Infect Control Hosp Epidemiol 2008;29:771-3. [Crossref] [PubMed]
- Lee JH, Kim J, Kim K, et al. Albumin and C-reactive protein have prognostic significance in patients with community-acquired pneumonia. J Crit Care 2011;26:287-94. [Crossref] [PubMed]
- Fernández-Ruiz M, Lopez-Medrano F, Varela-Pena P, et al. Monitoring of immunoglobulin levels identifies kidney transplant recipients at high risk of infection. Am J Transplant 2012;12:2763-73. [Crossref] [PubMed]
- Stevens RB, Mercer DF, Grant WJ, et al. Randomized trial of single-dose versus divided-dose rabbit anti-thymocyte globulin induction in renal transplantation: an interim report. Transplantation 2008;85:1391-9. [Crossref] [PubMed]
- Wong W, Agrawal N, Pascual M, et al. Comparison of two dosages of thymoglobulin used as a short-course for induction in kidney transplantation. Transpl Int 2006;19:629-35. [Crossref] [PubMed]
- Mourad G, Dussol B, Daugas E, et al. Is renal allograft dysfunction a risk factor for severe infection in kidney transplant recipients? Transplant Proc 2012;44:2821-3. [Crossref] [PubMed]
- Lau L, Kankanige Y, Rubinstein B, et al. Machine-Learning Algorithms Predict Graft Failure After Liver Transplantation. Transplantation 2017;101:e125-32. [Crossref] [PubMed]


