
An intronic polymorphism of NFATC1 gene shows a risk association with biopsy-proven acute rejection in renal transplant recipients
Introduction
Kidney transplantation has been considered as the optimal therapy for patients with end-stage renal diseases (1). Benefiting from novel immunosuppressive agents, incidence of acute rejection (AR) following kidney transplantation has dramatically declined over the past decades; however, approximately 10–15% of recipients currently suffer from AR during their first year post-transplant, which still adversely contribute to the loss of graft function (2-4). Recently, immunological factors, especially T cell-mediated immune responses, are believed to be strongly associated with the pathogenesis of AR episodes (5,6).
Nuclear factor of activated T cells (NFAT) was originally observed in the nuclear extracts from activated T cells and was reported to activate T cells by binding to the interleukin-2 (IL-2) promoter (7). Moreover, with regard to the extensive administration of cyclosporine A (CsA) and tacrolimus (TAC), which function by inhibiting calcineurin activity and NFAT nuclear translocation, as well as the consequent target gene transcription, the role of NFAT has been considered essential in the context of solid organ transplantation (8). The NFAT family of transcription factor consists of five members (NFATC1–NFATC5), which are all coded by different genes (9). In addition to NFATC5, which is activated in response to osmotic stress, all other NFAT proteins are regulated by the calcium and calcineurin signaling pathways (10-12). The calcium-related NFAT proteins consist of two conserved domains: (I) the Rel-homology region (RHR); and (II) the NFAT homology region (NHR), which is a regulatory domain containing a calcineurin-binding site that regulates calcium, as well as calcium and calcineurin signaling pathways (13,14). Moreover, the NHR is highly phosphorylated under steady-state condition, maintaining NFAT in an inactive state and restricted to the cytoplasm; NHR can be dephosphorylated under conditions of increased intracellular calcium levels and the activation of calcineurin, leading to NFAT cytoplasmic-nuclear trafficking (15,16). Once in the nucleus, NFAT proteins function as transcription factors by binding to their target promoter regions (12).
The NFATC1 proteins are encoded by NFATC1 gene and located on chromosome 18q23. In addition, mice lacking the NFATC1 protein develop splenomegaly associated with the hyperproliferation of both B and T cells, combined with altered levels of IL-4, IL-13, and granulocyte-monocyte colony stimulating factor (GM-CSF), indicating an essential role of NFATC1 on immune cell growth and transcription regulation (17). In solid organ transplantation, the regulation of gene expression (e.g., IL-2 and GM-CSF) by NFATC1 has been correlated with an over-immunosuppressive status, and is also observed in recipients who receive calcineurin inhibitors (CNIs) (18-20). While these studies illustrate the extensive relationship between NFATC1 proteins and the immune response, no studies have explored its influence on the AR episodes that occur following solid organ transplantation.
In this study, we performed a comprehensive analysis of NFATC1 single nuclear polymorphisms (SNPs) in renal transplant recipients using target sequencing (TS) based on next-generation sequencing (NGS), and 55 SNPs were detected. Then, 14 tagger SNPs were remained for further association analysis after the Hardy-Weinberg equilibrium (HWE) and major allele frequencies (MAF) analysis. The multivariable logistic regression analysis adjusted by two confounding clinical characteristics identified one significant SNP (rs2290154) with the remarkable pathological changes. Finally, our in vitro study transfected the mutated plasmid of rs2290154 and demonstrated that the mutation of rs2290154 promoted T cell proliferation by causing the increasingly translation of NFATC1 mRNA and synthesis of NFATC1 protein, which leads to the increased secretion of IL-2.
Methods
Ethics approval and consent to participate
Local ethics committee of the First Affiliated Hospital with Nanjing Medical University approved the protocols followed in this study (2016-SR-029). Written informed consents were obtained from all transplant recipients. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Recipients in this study was strictly limited to living-related transplantation of donors to lineal or collateral relatives not beyond the third degree of kinship or transplantation of kidney donors after cardiac death from 2011 to 2015.
Patients and immunosuppressive therapy
A total of 200 cases were selected among recipients who underwent kidney transplantation surgery between 1st February 2011 and 1st December 2015 at the renal transplant center of First Affiliated Hospital with Nanjing Medical University. Detailed medical records and materials, including age, gender, transplant date, duration of transplant, transplant times, immunosuppressive protocol, panel reactive antibody (PRA) and human leukocyte antigen (HLA) mismatch, were carefully extracted by two independent clinicians (Zijie Wang and Ruoyun Tan) for patient selection.
Detailed immunosuppressive protocol following renal transplant was presented in our previous study (21). Briefly, all recipients received a three- or four-drug immunosuppressive regimen consisting of CsA or TAC in combination with mycophenolate mofetil (MMF) and prednisone (Pred), with or without sirolimus. The dosage of CsA and TAC was started at 8 and 0.2 mg/kg/d, respectively, and adjusted according to the results of therapeutic drug monitoring of the serum creatinine levels. A dosage of 200 mg/d of intravenous methylprednisolone was adopted for each BPAR episode lasting 3 to 5 days. The BPAR diagnostic criteria were based on the comprehensive evaluation of histological and clinical characteristics. The clinical characteristics were comprised of a 20% increase in serum creatinine levels from baseline (not attributable to other cases), fever, urine protein, and pain in the region of the transplanted kidney; histologically, the BPAR diagnosis was confirmed by an allograft biopsy according to Banff 07 classification (22), and consequently, the i, g, t, v, and ptc scores for each case were collected, which represent the interstitial inflammation, glomerulitis, tubulitis, intimal arteritis, and peritubular capillaritis, respectively.
During the follow-up of 3 years post-transplant for each subject, recipients who experienced at least one BPAR episode were assigned to BPAR group, while recipients with stable allograft function (STA) during the follow-up were enrolled to stable control group.
Sample collection, preparation, and sequencing
The detailed procedures of sample collection and the TS steps were described in our previous study (23). In brief, peripheral blood samples (2 mL) were collected from each recipient included in our study and the DNA was extracted. Quantitative detection of the concentration and purity of genomic DNA (gDNA) was performed while the gene integrity was tested by agarose gel electrophoresis. Next, a pool containing upstream and downstream oligonucleotides specific to the targeted regions of interest was hybrids to the gDNA samples. Quantitative detection was performed and fragmentation was followed by end repair, dA tailing, and sequencing adaptor ligation with an ABI 9700 PCR instrument (ABI, USA). The adapter-ligated DNA was amplified by selective, limited-cycle PCR and then quantitatively analyzed. The prepared library (750 ng) was hybridized overnight and hybridized products were mixed with 200 µL MyOne Streptavidin T1 magnetic beads (Invitrogen, USA) for 30 min at room temperature. Mixture was amplified for 16 PCR cycles and quantitatively assessed. The captured libraries were denatured and loaded onto an Illumina cBot instrument at 12 to 16 pmol/L for cluster generation according to the manufacturer’s instructions. Up to 20 WUCaMP libraries were sequenced per HiSeq lane.
Data analysis
Sequencing data, including the number of altered chromosomes, genomic alternation information, and the depth of sequencing coverage were analyzed. All analyses were based on the human reference sequence UCSC build hg19 (NCBI build 37.2) using the Burrows-Wheeler Aligner (BWA) (24). Local alignment and duplication removal were completed by the application of the Genome Analysis Tool Kit (GATK) and Picard software. SNP detection was performed using dbSNP 132. Damaging or deleterious SNPs were predicted using Gemini software. Prediction tools, including sorting intolerant from tolerant (SIFT) and polymorphism phenotyping (PolyPhen), were used to analyze all human non-synonymous SNPs. In addition, putative somatic variant calls were detected with two separate programs (MuTect 1.1.5 and VarScan 2.3.6 software) by pairing each sample with its matched blood.
Cell culture
The Jurkat T cell was obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in 1640 medium supplemented with 10% fetal bovine serum (FBS, Gibco, Australia) and 1% penicillin-streptomycin in an incubator with humidified 5% CO2 at 37 °C. The plasmids pCDNA3.1, pCMV-NFATC1 and pCMV-NFATC1-S617 were synthesized by Genechem (Shanghai, China) and then the transfection was performed according to the manufacturer’s protocol. Detailed information of plasmids construction was presented in the Table S1. The cells were stimulated by PMA/Ionomycin mixture (Multi Sciences, Hangzhou, China) for 24 h among Stimulated group, pCDNA3.1 group, pCMV-NFATC1 group and pCMV-NFATC1-S617 group. The naive Jurkat T cell was defined as Control group.

Full table
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cultured cells by using RNA extraction kits (Tiangen, China) according to the manufacture’s protocol. Aliquots of total RNA were subjected to the RT reaction using PrimeScript RT reagent Kit (Takara, Japan). After reverse transcription, qRT-PCR were performed in triplicate in a 96-well plate containing 1 µL of synthesized cDNA, QuantiNovaTM SYBR Green PCR Kit (Qiagen, Germany) using StepOnePlusTM Real-Time PCR System (Applied Biosystems, USA) according to the manufacturer’s instructions. The following primers were used for qRT-PCR: NFATC1, Forward: 5'-CCCACTCCTAGTTGCCACAT-3', Reverse: 5'- CCTGCTTTCCTTCTTGTTGG-3'). Fold changes in mRNA expression were calculated using 2–ΔΔCt method and normalized based on GADPH with ABI Step One Software version 2.1. All experiment was repeated for at least three times.
Enzyme-linked immunosorbent assay (ELISA)
ELISA kits (Multi Sciences, Hangzhou, China) were used to determine the supernatant levels of IL-2 in each group at 24 h. The optical density (OD) value was measured at 450 nm on the microplate reader (Bio-Rad, USA). All procedures were followed strictly according to the manufacture’s protocols. All experiment was repeated for at least three times.
Cell proliferation assay
The cells were seeded into 96-well plates, at a density of 1×104 cells per well, for the cell proliferation assay. Cell proliferation was detected using CCK-8 (Dojindo Molecular Technologies Inc., USA) following to the manufacture’s protocols. The 10 µL CCK-8 was added to each well at the 24, 48, 72 and 96 h after stimulation, and then cultured for 4 h. The OD values were read at 450 nm on the microplate reader (Bio-Rad, USA). All experiment was repeated for at least three times.
Western blotting
The protein of cells was extracted using RIPA buffer (Beyotime, China) with PMSF, and quantified by the BCA Protein Assay Kit according to the manufacturer’s instructions (Beyotime, China). Protein samples were separated by 10% SDS-PAGE gels, and then transferred onto polyvinylidene fluoride (PVDF) membranes. Following blocking with 5% skimmed milk for 2 h at 24 °C, the PVDF membranes were incubated with rabbit antibody specific to human NFAT1 (#5861, Cell Signaling Technology, MA, USA, 1:1,000), or GAPDH (#5174, Cell Signaling Technology, MA, USA, 1:1,000), or FLAG (#14793, Cell Signaling Technology, MA, USA, 1:1,000) for overnight at 4 °C. The membranes were then incubated with anti-rabbit secondary antibody for 2 h at 24 °C (Thermo Fisher Scientific Inc., USA, 1:5,000). Chemiluminescence system (Bio-Rad, USA) and Image Lab Software (Bio-Rad Laboratories Inc., USA) were used to analyze the protein expression. All experiment was repeated for at least three times.
Statistical analysis
HWE and MAF analysis were performed using gene frequencies obtained by a single gene counting. We explored the linkage disequilibrium (LD) blocks and tagger SNPs using Haploview version 4.2 software. A genotype association analysis of tagger SNPs was performed using the dominant model (minor allele homozygotes plus heterozygotes vs. major allele homozygotes), recessive model (minor allele homozygotes vs. heterozygotes plus major homozygotes), additive model (major homozygotes vs. heterozygotes vs. minor homozygotes), HET model (major homozygotes vs. heterozygotes) and HOM model (major homozygotes vs. minor homozygotes). Comparisons of the genotypic frequencies between the STA and BPAR groups were assessed using a chi-square test. Genotypic distributions of tagger SNPs between two groups were compared with logistic regression model adjusted for confounding factors identified by general linear model (GLM). Then, significant tagger SNPs were included for further evaluation of the relationship with the pathological changes by using the multivariable logistic regression model. Odds ratios (OR) and 95% confidence intervals (95% CIs) were calculated using SPSS 13.0 software (SPSS Inc., Chicago, IL, USA). Significant SNPs with BPAR episodes and pathological changes in recipients were selected for the in vitro function validation. Results were presented as mean ± standard deviation (SD) from at least three independent experiments using GraphPad Prism 6.0 software (San Diego, CA, USA). Comparisons between mutation and wild-type groups were conducted using an unpaired Student’s t-test or 2-way ANOVA. A threshold of P<0.05 was considered significant. The Bonferroni correction method was performed to avoid inflation of P value when multiple comparisons happened.
Results
Study population
The baseline characteristics of two study groups are presented in Table 1. No significant differences associated with the donor age, donor gender, recipient age, and recipient gender were observed (P>0.05). Among all the enrolled recipients, the majority (88.5%) received donor grafts from a donor after cardiac death (DCD). Although the mean levels of pre-transplant PRA and HLA mismatches, as well as the sirolimus usage proportion in the BPAR group were relatively higher than those in the STA group, no statistical differences were found (P>0.05).
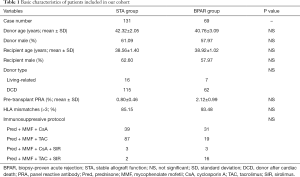
Full table
SNP identification
A total of 55 SNPs located in the NFATC1 gene were identified by TS based on NGS in our study. The genetic information of all detected SNPs was presented in Table S2. After HWE and MAF analysis, as well as the removal of SNPs with HWE less than 0.05 and/or minor allele frequencies (MAF <0.05), 20 SNPs were obtained (Table S3). Furthermore, 14 tagger SNPs were selected for further association analysis by LD analysis (Figure 1).
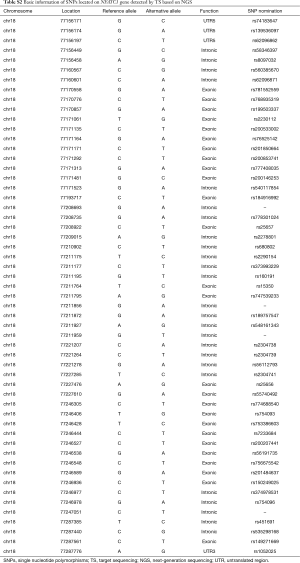
Full table
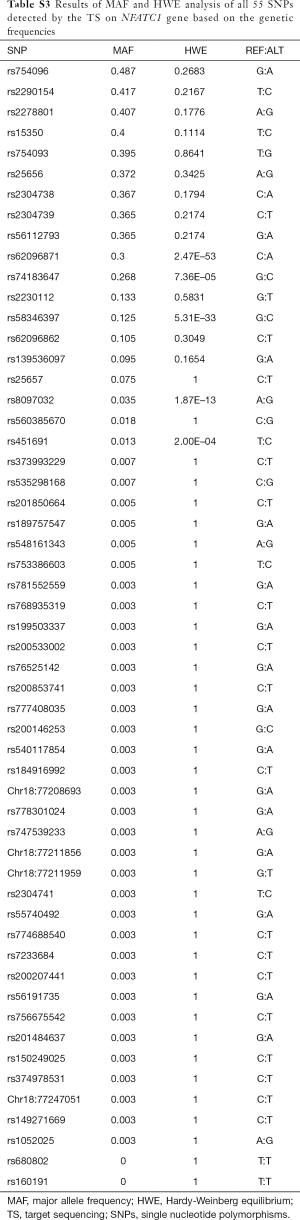
Full table
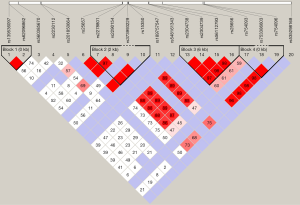
Association of NFATC1 SNPs with AR episodes
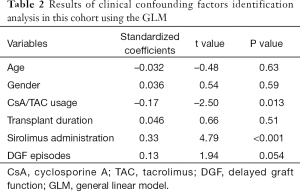
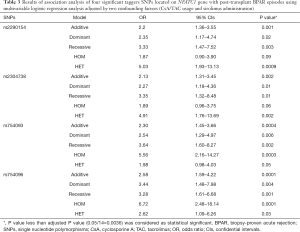
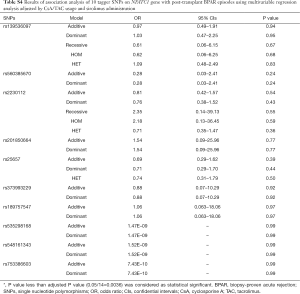
Next, GLM analysis identified two confounding factors, CsA/TAC usage and sirolimus administration (CsA/TAC usage: t=–2.50, P=0.013; sirolimus administration: t=4.79, P<0.001; Table 2). Then, a logistic regression analysis with an adjustment for CsA/TAC usage and sirolimus administration was performed for all five analytical models (dominant, recessive, additive, HET and HOM) to explore the relationship between NFATC1 SNPs and BPAR episodes following kidney transplantation. Overall, four SNPs in the NFATC1 gene (rs2290154, rs2304738, rs754093 and rs754096) were summarized to be statistically significant between STA and BPAR groups (Table 3). We did not observe any significant difference in other tagger SNPs (Table S4). In addition, four haplotypes were identified by LD analysis and there was no significant difference with BPAR episodes.
Full table
Full table
Full table
Association of NFATC1 SNPs with pathological examination
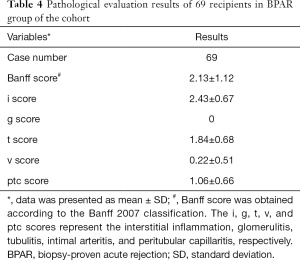
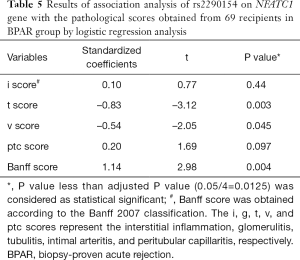
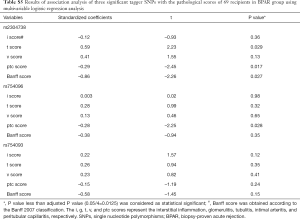
We examined all 69 recipients suffering from the BPAR episodes and reviewed the each case of pathological slides by two independent pathological professors (H Chen and RY Tan) according to Banff 07 criteria. The results of pathological evaluation were presented in Table 4. We explored the relationship between four significant SNPs and pathological changes, and found that only one SNP, rs2290154, was significantly associated with Banff score and t score using multivariable logistic regression (Banff score: t=–3.12, P=0.003; t score: t=2.98, P=0.004; Table 5). However, no difference of other three SNPs (rs2304738, rs754093, and rs754096) was observed (Table S5).
Full table
Full table
Full table
NFATC1 rs2290154 mutant promotes T cells proliferation and activation
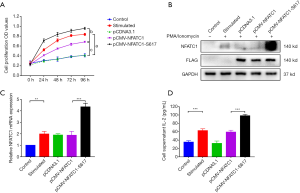
To identify the biological function of rs2290154 mutation on the NFATC1 gene, we transfected the wild-type and mutated plasmids of rs2290154 into T lymphocytes, respectively. We tested the cell proliferation and found that NFATC1 rs2290154 mutant significantly promoted the proliferation of T lymphocytes, especially at 48 and 72 h after stimulation (Figure 2A). Then, we examined the translation of NFATC1 mRNA and expression of NFATC1 protein, the results showed that NFATC1 mRNA and NFATC1 protein was significantly increased by the NFATC1 rs2290154 mutant in T lymphocytes (Figure 2B,C), along with the remarkable increase of IL-2 in supernatant (Figure 2D).
Discussion
Renal transplant recipients are generally exposed to combined immunosuppressive therapy with a limited therapy window, which is associated with a higher rate of BPAR episodes. The NFATC1 gene was found to influence the susceptibility to AR by altering the activity and expression of NFATC1 protein, which plays an important role in gene transcription for mounting an immune response (25). Here, we conducted an association study of Chinese renal transplant recipients to comprehensively explore the effects of the NFATC1 gene on the pathogenesis of AR episodes. Sequencing study has identified one SNP, rs2290154, was significantly associated with the BPAR episodes and pathological changes, especially in the Banff score and renal tubulitis. Furthermore, our in vitro study has identified what we believe is a novel molecular mechanism by which NFATC1 rs2290154 mutant promoted the T lymphocytes activation and proliferation through increasing the translation of NFATC1 mRNA and expression of NFATC1 protein, and the secretion of IL-2, which is responsible for the increased risk of AR episodes following renal transplant.
It is well established that NFAT transcription factors play critical roles in CD4 T cell activation and differentiation and calcineurin/NFAT network has an important function in the generation and function of activated T lymphocytes, including CD4+ and CD8+ T cells (26,27). In one study, a defective nuclear translocation of NFATC1 has been described for NFATC1 in CD8+ T cells, not in CD4+ T cells (27). Consistently, ablation of NFATC1 results in an impaired formation of F-actin rings around the immunological synapse in cytotoxicity T lymphocyte (CTLs) upon TCR stimulation, indicating that NFATC1 controls the effector function of CD8+ T cells at multiple levels (26). To date, no association studies have been performed to explore the relationship between NFATC1 gene polymorphisms and the AR episodes that occur following kidney transplantation. Moreover, the mechanisms of such interactions remain unclear. In a clinical study performed in cases of human allograft rejection, an increase in calpain was observed in infiltrating T cells, providing novel prospects for pharmacological manipulation of the calpain/calpastatin balance during solid organ transplantation (28). Since calpain is a calcium-activated protease which is involved in the activation of NF-κB and NFAT, the outcomes in this study indicate that the NFAT gene may indirectly participate in the development of rejection. In our study, we sequenced the loci mutations on NFACT1 gene and identified one SNP mutant may have a positive impact on the NFATC1 protein expression, as well as the cell proliferation, explaining the results observed in our cohort. We provided the genetic evidence of NFATC1 in the pathogenesis of AR episodes following renal transplant, which could be used as a candidate gene for the regulation of massive T lymphocyte activation and proliferation.
Recently, published GWAS studies summarized that nearly 80% of trait-associated SNPs are located in non-coding regions, such as intron and untranslated region (UTR) (29). Results from numerous studies have implicated crucial regulatory functions to noncoding intronic loci on human genes (30,31). From our sequencing data and in vitro results, we have concluded that one intron SNP located in NFATC1, rs2290154, may contribute to the significant proliferation and activation of T lymphocytes in AR episodes by the mutation from T allele to C allele. Since no previous studies have associated this mutation with the function of T cells, we were unable to explain the detailed regulatory mechanism of rs2290154 mutant in AR episodes. Furthermore, we have simultaneously verified that two SNPs, rs25657 and rs2278801, in linkage with rs2290154 were not in the chromatin regions annotated as promoters and enhancers. Taken together, our results suggest that the NFATC1 gene may be a plausible candidate gene for the pathogenesis of post-transplantation AR episodes by encoding a transcription factor (rs2290154 mutant) which regulates gene transcription during an immune response (32).
Some limitations associated with our study should also be acknowledged: (I) this study contained a limited number of subjects from a single transplant center, which inevitably led to the limited power to detect a significant association. Thus, the results needs to be validated by replication in different patient cohorts; (II) there was a lack of information pertaining to the exposure to environmental factors in this study (e.g., diet and lifestyle); (III) our in vitro study reveals the positive regulation of NFATC1 rs2290154 mutant on T cells proliferation, which partly explained the results observed in the occurrence of BPAR episodes of this cohort; and (IV) with regard to the complexity of AR episodes following kidney transplantation, various genetic and non-genetic factors have been identified to partially contribute to the BPAR; while in our study, mainly the genetic factors were explored with the lack of non-genetic factors investigation, which should be interpreted with great caution. However, extensive explore to the influence of this mutant on the T cell activation, differentiation and metabolism are still warrant.
In summary, our study firstly reported the crucial function of NFATC1 gene in the occurrence of AR episodes. Using the TS based on the NGS technology, we have identified one significant SNP, rs2290154, is significantly associated with the remarkable renal allograft pathological changes. Our in vitro study further observed that the NFATC1 rs2290154 mutant could significantly promoted the T cells proliferation, increased the translation of NFATC1 mRNA and the expression of NFATC1 protein, as well as the secretion of IL-2. Given the known trigger function of NFATC1 in T lymphocytes, these findings suggest a possible link between genetic variation in NFATC1 and the onset of AR following renal transplant. Future studies will be required to validate the importance of NFATC1 rs2290154 mutant in the progression of AR episodes.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (grant numbers 81900684, 81870512, 81770751, 81570676, 81470981, 81100532), Project of Jiangsu Province for Important Medical Talent (grant number ZDRCA2016025), the “333 High Level Talents Project” in Jiangsu Province (grant numbers BRA2017532, BRA2016514, BRA2015469), the Standardized Diagnosis and Treatment Research Program of Key Diseases in Jiangsu Province (grant number BE2016791), the Open Project Program of Health Department of Jiangsu Province (grant number JSY-2-2016-099), Jiangsu Province Natural Science Foundation Program (grant number BK20191063).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Local ethics committee of the First Affiliated Hospital with Nanjing Medical University approved the protocols followed in this study (2016-SR-029). Written informed consents were obtained from all transplant recipients. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang Z, Han Z, Tao J, et al. Transforming growth factor-β1 induces endothelial-to-mesenchymal transition via Akt signaling pathway in renal transplant recipients with chronic allograft dysfunction. Ann Transplant 2016;21:775-83. [Crossref] [PubMed]
- Ekberg H, Tedesco-Silva H, Demirbas A, et al. Reduced exposure to calcineurin inhibitors in renal transplantation. N Engl J Med 2007;357:2562-75. [Crossref] [PubMed]
- Cole EH, Johnston O, Rose CL, et al. Impact of acute rejection and new-onset diabetes on long-term transplant graft and patient survival. Clin J Am Soc Nephrol 2008;3:814-21. [Crossref] [PubMed]
- Dunn TB, Noreen H, Gillingham K, et al. Revisiting traditional risk factors for rejection and graft loss after kidney transplantation. Am J Transplant 2011;11:2132-43. [Crossref] [PubMed]
- Ayed K, Abdallah TB, Bardi R, et al. Plasma levels of soluble CD30 in kidney graft recipients as predictors of acute allograft rejection. Transplant Proc 2006;38:2300-2. [Crossref] [PubMed]
- Hariharan S. Long-term kidney transplant survival. Am J Kidney Dis 2001;38:S44-50. [Crossref] [PubMed]
- Shaw JP, Utz PJ, Durand DB, et al. Identification of a putative regulator of early T cell activation genes. Science 1988;241:202-5. [Crossref] [PubMed]
- Liu J, Farmer JD Jr, Lane WS, et al. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 1991;66:807-15. [Crossref] [PubMed]
- Xu Q, Qiu X, Jiao Z, et al. NFATC1 genotypes affect acute rejection and long-term graft function in cyclosporine-treated renal transplant recipients. Pharmacogenomics 2017;18:381-92. [Crossref] [PubMed]
- López-Rodríguez C, Aramburu J, Jin L, et al. Bridging the NFAT and NF-kappaB families: NFAT5 dimerization regulates cytokine gene transcription in response to osmotic stress. Immunity 2001;15:47-58. [PubMed]
- Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol 1997;15:707-47. [Crossref] [PubMed]
- Hogan PG, Chen L, Nardone J, et al. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev 2003;17:2205-32. [Crossref] [PubMed]
- Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol 2005;5:472-84. [Crossref] [PubMed]
- Lopez-Rodríguez C, Aramburu J, Rakeman AS, et al. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc Natl Acad Sci U S A 1999;96:7214-9. [Crossref] [PubMed]
- Shibasaki F, Price ER, Milan D, et al. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature 1996;382:370-3. [Crossref] [PubMed]
- Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature 1992;357:695-7. [Crossref] [PubMed]
- Hodge MR, Ranger AM, Charles de la Brousse F, et al. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity 1996;4:397-405. [Crossref] [PubMed]
- Zahn A, Schott N, Hinz U, et al. Immunomonitoring of nuclear factor of activated T cells-regulated gene expression: the first clinical trial in liver allograft recipients. Liver Transpl 2011;17:466-73. [Crossref] [PubMed]
- Sommerer C, Zeier M, Meuer S, et al. Individualized monitoring of nuclear factor of activated T cells-regulated gene expression in FK506-treated kidney transplant recipients. Transplantation 2010;89:1417-23. [Crossref] [PubMed]
- Sommerer C, Zeier M, Czock D, et al. Pharmacodynamic disparities in tacrolimus-treated patients developing cytomegalus virus viremia. Ther Drug Monit 2011;33:373-9. [Crossref] [PubMed]
- Wang Z, Zheng M, Yang H, et al. Association of genetic variants in CYP3A4, CYP3A5, CYP2C8, and CYP2C19 with tacrolimus pharmacokinetics in renal transplant recipients. Curr Drug Metab 2019;20:609-18. [Crossref] [PubMed]
- Solez K, Colvin RB, Racusen LC, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant 2008;8:753-60. [Crossref] [PubMed]
- Wang Z, Yang H, Si S, et al. Polymorphisms of nucleotide factor of activated T cells cytoplasmic 2 and 4 and the risk of acute rejection following kidney transplantation. World J Urol 2018;36:111-6. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- Peng SL, Gerth AJ, Ranger AM, et al. NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity 2001;14:13-20. [Crossref] [PubMed]
- Klein-Hessling S, Muhammad K, Klein M, et al. NFATc1 controls the cytotoxicity of CD8+ T cells. Nat Commun 2017;8:511. [Crossref] [PubMed]
- Leung-Theung-Long S, Mondor I, Guiraud M, et al. Impaired NFAT transcriptional activity in antigen-stimulated CD8 T cells linked to defective phosphorylation of NFAT transactivation domain. J Immunol 2009;182:6807-14. [Crossref] [PubMed]
- Letavernier E, Dansou B, Lochner M, et al. Critical role of the calpain/calpastatin balance in acute allograft rejection. Eur J Immunol 2011;41:473-84. [Crossref] [PubMed]
- Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 2009;106:9362-7. [Crossref] [PubMed]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57-74. [Crossref] [PubMed]
- Gerstein MB, Kundaje A, Hariharan M, et al. Architecture of the human regulatory network derived from ENCODE data. Nature 2012;489:91-100. [Crossref] [PubMed]
- Horsley V, Pavlath GK. NFAT: ubiquitous regulator of cell differentiation and adaptation. J Cell Biol 2002;156:771-4. [Crossref] [PubMed]


