
Identification of hub genes and potential molecular mechanisms of chickpea isoflavones on MCF-7 breast cancer cells by integrated bioinformatics analysis
Introduction
As of 2018, breast, lung, and colorectal cancers were estimated to be the three most common types of cancer, accounting for one-half of all cancer cases in women, with breast cancer alone considered to constitute 30% of all new diagnoses of cancer in women (1). According to the American Society of Clinical Oncology, approximately 60% to 75% of women with breast cancer is estrogen and progesterone receptor-positive (2). Furthermore, as reported by epidemiological studies, the prevalence of breast cancer is generally lower in Asian than in North American and European countries (3). However, the incidence of breast cancer in Asians increased when Asians move to live in other regions, and almost equaled the rates of the host country recently (4). Of late, the preventative effects of phytoestrogens on breast cancer have attracted significant attention and have become a major focus of breast cancer research.
Isoflavones are a class of phytoestrogens that are structurally similar to mammalian estrogens. Epidemiological studies have suggested that higher consumption of legumes containing large amounts of phytoestrogen might contribute to the lower incidence in Asia of some cancers like breast cancer, colon cancer, and colorectal cancer (5). Chickpea (Cicer arietinum L.) is an important global legume crop. Germination increases the phenolic content of seeds, and particularly in chickpea, the isoflavone content is increased by over 100 fold, mainly because of the increase of formononetin and biochanin (6). According to our previous study, chickpea isoflavones could not only significantly inhibit the survival, proliferation, and migration of MCF-7 cells but could also induce their apoptosis (7,8). In order to explore the mechanism of isoflavones inhibiting the formation and development of breast carcinoma, we used the transcriptome sequencing of Michigan Cancer Foundation-7 (MCF-7) cell treatment with isoflavones to analyze the differentially expressed genes, and clarify their biological functions. Moreover, the mechanism behind the inhibitory effect of chickpea isoflavones on the proliferation of human breast cancer cells was deduced, yielding data buttressing a theoretical basis for explaining how human breast cancer can be treated with isoflavones, while offering possible gene targets for the development of new anti-cancer drugs for use in breast cancer.
Methods
Cell culture
MCF-7 cells were provided by the Chinese Type Culture Collection, CAS (Shanghai, China). Cells were grown in Dulbecco’s modified Eagle medium (DMEM) with 4.5 g/L glucose and 0.37% sodium bicarbonate (Gibco, Rockville, MD, USA), and a fully humidified incubator (Binder, Germany) at 37 °C with 95% air and 5% CO2.
Antiproliferation assay
The MCF-7 cells in the logarithmic growth phase were digested with 0.25% trypsin digest and separately seeded on 96-well plates at a density of 5×103 cells per well. After 24 hours in the incubator, the cells were mixed with chickpea isoflavones to a final concentration of 10, 20, 30, 40, 50, and 60 µg/mL for 24, 48, and 72 hours, respectively. Five replicates were set for each concentration. Moreover, an equal volume of dimethyl sulfoxide (DMSO) was added to the control group. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used to determine the antiproliferative effects of the cells. The optical density value of each well was measured at 570 nm. The experiment was repeated 3 times. The IC50 calculator was used to simulate the integration curve, and the half-inhibitory concentration of chickpea isoflavones on MCF-7 cells was calculated. Apoptosis was quantified by using Annexin V-FITC (Signalway Antibody) /PI staining and flow cytometer (Becton Dickinson, NJ, USA).
Cell morphology observation
For test sample treatments, chickpea isoflavones were added to the culture media to a final concentration of 32.8 µg/mL in DMSO for 48 hours; control samples were treated with an equal volume of DMSO only (9). Each group had 3 replicate samples prepared. The morphological changes of cells were observed by inverted microscope.
RNA extraction and library preparation
Total RNA was extracted using the TRIzol Kit (ComWin Biotech, Beijing, China) from control and test cells according to the manufacturer’s instructions. Then, the total RNA was treated with RNase-free DNase I (Takara, Dalian, China) for 30 min at 37 °C to remove residual DNA. The RNA quality was verified by RNase-free agarose gel electrophoresis, and the total RNA concentration was measured at 260 and 280 nm. RNA samples with 260/280 nm ratios between 1.8 and 2.0 were used for subsequent analyses.
The transcriptome assembly library was constructed by RNA from the above samples. Briefly, total mRNA was enriched by Oligo (dT) beads. Then, the enriched mRNA was fragmented into short fragments using fragmentation buffer and reverse transcripted into cDNA with random primers. Second-strand cDNA was synthesized by DNA polymerase I, RNase H, dNTP, and buffer. Next, the cDNA fragments were purified with QiaQuick PCR extraction kit, end-repaired, poly(A)-added, and ligated to Illumina sequencing adapters. The ligation products were size-selected by agarose gel electrophoresis, PCR-amplified, and sequenced using Illumina HiSeqTM 2500 by Gene Denovo Biotechnology Co. (Guangzhou, China).
Transcript quantification and differential expression analysis
The original sequencing data were sequenced to obtain high-quality clean reads through the removal of joint and low-quality reads, and the transcript abundances in the RNA-sequencing (RNA-Seq) samples were quantified using Kallisto version 0.44 (https://pachterlab.github.io/kallisto/about.html). Kallisto performs a pseudo-alignment using the reference transcriptome to determine the compatibility of reads with target transcripts. After indexing the human reference transcriptome (mRNA: human ensemble cDNAs; LncRNA: human LncRNAs of NONCODE v.5.0), the previously trimmed paired-end reads were mapped to the indexed transcriptome. The differential expression was analyzed by edge R. We set the cut-off criteria as |logFC| >1, with the P value <0.05 to screen the differentially expressed genes/LncRNA (DEGs/DEL).
Functional analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/) analyses are used for annotating genes and gene products, systematically analyzing gene functions, and other purposes (10-12). WEB-based Gene SeT Analysis Toolkit (WebGestalt) is a functional enrichment analysis web tool. In order to analyze the DEGs at the functional level, GO enrichment and KEGG pathway analysis were performed using the WebGestalt online tool (http://www.webgestalt.org/option.php). The criteria were set as follows: the minimum number of genes for a category was 5, and the maximum number of genes for a category was 2,000. Moreover, BH method (Benja-mini-Hochberg FDR) was performed for multiple test adjustment. A P value <0.05 was considered statistically significant.
Protein-protein interaction (PPI) network construction
We used the online databases, STRINGdb, and Gephi (https://gephi.org/), to construct the PPI network of DEGs and analyzed the subnet with the MCODE toolkit based on the METASCAPE website (http://metascape.org/gp/index.html). Then, a co-expression network of the top 10 long non-coding RNAs (lncRNAs) and messenger RNAs (mRNAs) was constructed based on Pearson’s correlation with a coefficient >0.95. The PPI network of co-expressed genes of the top 10 lncRNAs was constructed from the network analyst website (https://www.networkanalyst.ca/faces/home.xhtml).
Primer design and validation
To confirm the accuracy of the clustering relationship provided by the PPI network, 19 genes were selected for quantitative PCR validation. Primers of the candidate genes are shown in Table 1. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method was completed.
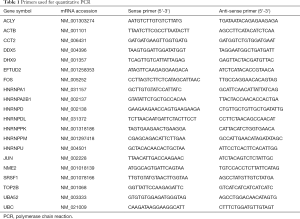
Full table
Survival analysis
We employed The Cancer Genome Atlas (TCGA) data to perform the survival analysis, which contained 1,075 breast cancer samples with the high degree hub genes as input. Kaplan-Meier survival analysis was used to compare the survival time of the different groups. A P value <0.05 was considered to indicate a statistically significant difference. Moreover, the deregulation of the expression of these genes was demonstrated by the immunohistochemistry staining obtained from the Human Protein Atlas database.
Results
Antiproliferative effects of chickpea isoflavones on MCF-7 cells
The antiproliferative effects of various concentrations (10 to 60 µg/mL) of chickpea isoflavones on the human breast cancer cell lines MCF-7 were assessed using the MTT assay. Chickpea isoflavones dramatically inhibited the proliferation of MCF-7 cells in a time-dependent and dose-dependent manner (Figure 1A). The IC50 calculator was used to simulate the integration curve, and the 48-hour half-inhibitory concentration of chickpea isoflavones on MCF-7 cells was calculated to be 32.8 µg/mL.
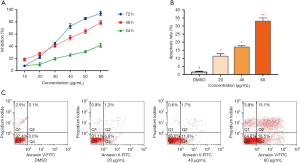
Flow cytometry was used to detect the effect of different doses of MCF-7 cells on apoptosis. The results of the analysis showed that the apoptosis rate had a dose-effect relationship with the treatment dose, and at a 60 µg/mL dose, the highest apoptosis rate was significantly different from that of the control group (P<0.01, Figure 1B,C).
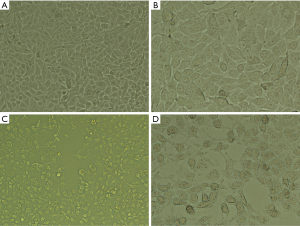
Morphological changes of the cells treated with chickpea isoflavones in MCF-7 cells
Chickpea isoflavones had an obvious inhibitory effect on the proliferation of MCF-7 cells (Figure 2). In the control group (DMSO), human breast cancer cells showed full adherent growth, compact arrangement, basically uniform size, regular shape, large volume, uniform distribution of chromatin in the nucleus, clear nucleoli, large and complete nucleus, and center or edge migration. After being treated with chickpea isoflavones for 48 hours, the MCF-7 cells were mostly blurred in contour, shrunken to a round shape, and had reduced cell density, reduced cell volume, increased space, different cell sizes, irregular morphology, concentrated chromatin, vacuolar like changes in the cytoplasm, and even “sprouting”.
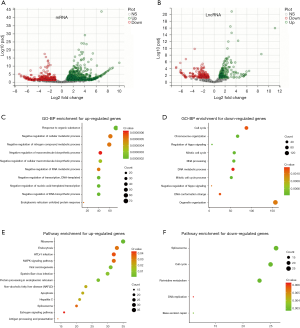
Identification of the differentially expressed transcripts (DETs) and function enrichment
A total of 1,094 DETs were obtained, including 729 up-regulated transcripts (482 genes) and 365 down-regulated transcripts (296 genes). Meanwhile, there were 378 differentially expressed long non-coding RNAs (lncRNAs) obtained, 204 of which were up-regulated, and 174 of which were down-regulated. Figure 3A,B shows the DEGs/DEL using a volcano plot. To further explore the biological processes (BPs) and pathways in which the overlapped genes were involved, all DEGs were uploaded to WebGestalt, and the results were obtained. GO analysis suggested that the up-regulated DEGs were significantly enhanced by BPs, including organic substances, cellular metabolism, and nitrogen compound metabolism (Figure 3C). However, the down-regulated DEGs were enhanced by organelle organization, cell cycles, and DNA metabolic processes (Figure 3D). As for biological pathways, the KEGG pathways analysis showed that the up-regulated DEGs were enriched with viral carcinogenesis, MAPK signaling pathways, HTLV-I infections, endocytoses, and ribosomes (Figure 3E), while the down-regulated DEGs were enriched with pyrimidine metabolism, cell cycles, and spliceosomes (Figure 3F).
PPI network construction
First, we constructed PPI networks of the up-regulated genes and down-regulated genes using software from the Stringdb and Gephi online databases. A total of 11 nodes (4 hub genes up-regulated; 7 hub genes down-regulated) with degrees >10 were chosen in up-regulated and down-regulated networks, respectively (Figure 4A). Secondly, based on Pearson’s correlation and Gephi software, we further constructed the co-expression networks of the top 10 up-regulated and down-regulated lncRNAs and co-expressed genes. The MCODE analysis showed that in the top 10 up-regulated lncRNAs, 4 nodes with a correlation coefficient >0.95 were positively correlated with co-expressed genes, which were involved in the peptide chain elongation. Meanwhile, in the top 10 down-regulated lncRNAs, only 1 node with a correlation coefficient >0.95 was positively correlated with co-expressed genes in the mRNA splicing process (Figure 4B). Finally, we constructed a PPI network of lncRNA co-expression genes based on the network analyst website. The results showed a PPI network with the interaction between 8 HNRNP family and 5 apoptosis-related genes via ubiquitin C (UBC), which indicated that the cause of isoflavone-induced apoptosis might be related to the negative regulation of the HNRNP family gene, as shown in Figure 4C.
Validation of RNA-Seq results by quantitative PCR
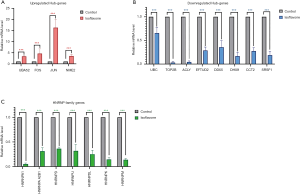
According to the clustering relationship provided by the PPI network, we further verified the RNA-Seq results of 19 genes by quantitative PCR, including HNRNP family genes, up-regulated hub genes and down-regulated hub genes in the control group and test group that had chickpea isoflavones. We observed the same expression patterns as RNA-Seq analysis with statistical significance (Figure 5).
Survival analysis of the hub-genes
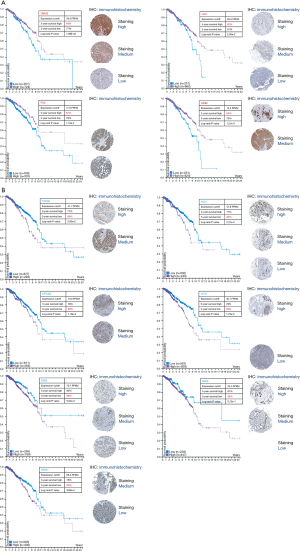
In order to further clarify whether isoflavone-induced abnormal expression of hub genes has a positive effect on the overall survival of breast cancer patients, we conducted survival analysis using the Human Protein Atlas (HPA) to evaluate the relationship between the hub genes of the PPI network and overall survival (OS). We found that high expression of 4 hub genes (UBA52, FOS, JUN, NME2) in the up-regulated genes could improve the survival of patients, and the low expression of 7 hub genes (TOP2B, ACLY, EFTUD2, DDX5, DHX9, CCT2, SRSF1) in the down-regulated genes could significantly improve the survival of patients; all Kaplan-Meier plots were calculated according to the critical optimal cut-off value of each gene provided by HPA, and a P value by the Log Rank Test less than 0.05 was considered statistically significant (Figure 6). Moreover, we integrated the immunohistochemistry results of each gene in HPA on the survival curve.
Discussion
Cancer is a global public health problem and is one of the primary causes of death worldwide. Isoflavone phytoestrogens show anti-cancer activity and have attracted attention from many researchers around the world. Their antitumor mechanism includes inhibiting the activity of tyrosine-protein kinase (PTK) (13) and topoisomerase II (14), controlling the cell cycle (15), and inducing apoptosis (16,17). In recent years, drugs that induce the apoptosis of cancer cells have become increasingly popular. It has been discovered that hummus isoflavone has two different regulation functions for breast cancer cell proliferation. A low concentration of isoflavone has a significant estrogenic effect while a high concentration has an inhibiting effect on the growth of breast cancer cells (7). To explore the molecular mechanism of chickpea isoflavones inhibiting breast cancer cell proliferation, we used the RNA-seq method to analyze the changes of gene expression in breast cancer MCF-7 cells after admixture with chickpea isoflavones.
Soy isoflavone is a kind of natural estrogen and a secondary metabolite produced during the process of soybean growth, including daidzein, genistein, daidzin, and genistein. The major compounds in chickpea seeds are formononetin, biochanin A, ononin, and sissotrin (9). The effect of germination on bioactive components in terms of the antioxidant capacity and total isoflavone content in legume seeds was previously investigated; germination increased the total isoflavone content and antioxidant capacity of most seeds, particularly in chickpea seeds (6). The nutritional value of chickpeas is high, and chickpeas are a good source of carbohydrates and protein compared to other legumes, which account for about 80% of the total quality of dried seeds (18,19). Chickpeas are cholesterol-free and are a good source of dietary fiber, vitamins, and minerals (20,21). They are widely cultivated in western Asia and the Near East and are currently grown in more than 50 countries in western and southern Asia, North Africa, the Middle East, Southern Europe, the Americas, and Australia. Globally, chickpeas are the third largest producer of legumes after soybeans and peas (22).
Chickpea isoflavones do cause apoptosis, activate the estrogen signaling pathway, and increase protein synthesis, but also down-regulate the variable shear of mRNA. In our research, isoflavone inhibited the expression of HNRNP family genes, which is a phenomenon that should be further explored. Heterogeneous nuclear ribonucleoprotein (HNRNP) is a superfamily of proteins. According to different physiological functions, the member can be divided into two categories. The first is characterized by being located in the nucleus, and these cannot be shuttled between the nucleus and cytoplasm. Proteins, such as HNRNP B, C, etc., bind to precursor mRNA to form a complex, which detaches from the complex before the precursor mRNA is transported out of the nucleus. The second type of protein can shuttle freely between the nucleus and the cytoplasm. For example, hnRNP A, D, E, I, K, which form a complex with the precursor mRNA, are accompanied by the entry of precursor mRNA into the cytoplasm through the nuclear pore complex, and complete the precursor with other proteins like RNA polymerase II. The shear processing and modification of the mRNA are then dissociated and returned to the nucleus, ready to participate in the next round of transport (23). A series of experiments in vitro have shown that HNRNPD (24), HNRNP A2/B1 (25), and HNRNP A1 (26) can bind to the telomere DNA. The results showed that the length of telomeres in CB3 cells that did not express HNRNP A1 was significantly shorter than that in CB7 cells expressing HNRNP A1 (27). In the Hela cells, the expression of HNRNP A1 /A2 /B1 gene was reduced by RNA interference, and the length of the G-tail of the back-end grain changed significantly (28). Current research shows that the down-regulation and knock-out of the HNRNP family gene inhibits the occurrence of apoptosis. For example, knockdown of HNRNP A2/B1 inhibits cell proliferation, invasion, and cell-cycle-triggering apoptosis in cervical cancer via PI3K/AKT signaling pathway (29), while HNRNP K plays a protective role in apoptosis in podocytes induced by TNF-α (30). HNRNP protein was originally discovered as a regulator of alternative splicing: the process of controlling the removal of introns and the selective linking of exons, through which multiple transcripts and subsequent proteins can be expressed from 1 gene. Alternative splicing affects genes in all key cell processes, including apoptosis. In the process of cancer growth, damaged apoptosis control is helpful for the survival of cells carrying molecular distortion and for their unrestricted proliferation and chemical resistance (31). Ubiquitination is a process that has been associated with protein degradation, DNA repair, cell cycle regulation, kinase modification, endocytosis, and regulation of other cell signaling pathways. Ubiquitin is highly expressed in various tumor tissues, and the down-regulation of ubiquitin molecular expression can inhibit lung cancer cell line proliferation (32). Ubiquitin C (UBC) is one of the four genes encoding ubiquitin in mammalian genome. It is described as the gene most responsive to cell therapy such as ultraviolet radiation, heat shock, oxidative stress and translation damage. Although the association between HNRNP and UBC facilitated by the PPI network is obvious, further research is still needed to consider the extensive biological function of UBC and how it connects to isoflavone, HNRNP, and the apoptosis pathway.
Although the survival analysis was provided by HPA, the sample data are all quantitative results of RNA-Seq, the optimal cut off value of each gene is fragments per kilobase of transcript per million mapped reads (FPKM), and each Kaplan-Meier (KM)-plot distinguishes high and low risk groups based on this optimal cut-off value. Even so, we found that not every hub gene could be used as a risk factor for breast cancer prediction. Among the up-regulated hub genes, only the up-regulated FOS gene could significantly improve the 5-year survival period. The FOS gene encodes leucine zipper proteins that can dimerize with proteins of the JUN family, thereby forming the transcription factor complex AP-1 (33). The FOS proteins have been implicated in the regulation of cell proliferation, differentiation, and transformation. In some cases, the expression of the FOS gene has also been associated with apoptotic cell death. In addition, NME2 also plays critical roles in many aspects of cancer biology, including cancer cell proliferation, survival, apoptosis, drug resistance, and metastasis (34). Moreover, the UBA52 gene encodes a fusion protein consisting of ubiquitin at the N terminus and ribosomal protein L40 at the C terminus (35). The major role of ubiquitin is to target cellular proteins for degradation by the 26S proteosome, and it is involved in the maintenance of chromatin structure, the regulation of gene expression, and stress response (36). In the down-regulated hub genes, we found that the down-regulation of DDX5, DHX9, and CCT2 significantly contributed to improving the survival time of breast cancer patients. DDX5 has an important role in transcription regulation with multiple, sequence-specific transcription factors but has also been implicated in other processes such as microRNA biology, RNA splicing, and ribosome biogenesis (37). DDX5 is frequently overexpressed in breast cancer (38), functions as a transcriptional coactivator for E2F1 to promote the expression of genes required for cell proliferation, and E2F1 is also frequently amplified and overexpressed in breast cancer (39). DDX5 knockdown in breast cancer cells with the E2F1 gene amplification blocked cancer cell proliferation and resulted in down-regulated expression of DNA replication factors. DHX9 is a DExH-box helicase family member, which participates in multiple levels of gene regulation, including DNA replication, transcription, translation, RNA transport, and microRNA processing. It has been implicated in tumorigenesis and DHX9 suppression, eliciting an apoptotic or senescence response by activating p53 signaling (40). CCT2 is a molecular chaperone that is a member of the chaperonin containing TCP1 complex (CCT), also known as the TCP1 ring complex (TRiC), and it helps other proteins to properly assemble and fold while mediating misfolded protein degradation. CCT2 and TCP1, as members of the TriC chaperone complex, are essential for the growth and survival of breast cancer cells and are closely related to cancer-driven genes such as P13K (41). CCT2 is highly expressed in triple-negative breast cancer cell lines such as MDA-MB-231, and the increased CCT protein in breast cancer cells underlies the cytotoxicity of CT20p. CCT is a potential target for therapeutic intervention for breast cancer treatment (42). SRSF1 forms an anti-apoptotic form of transcripts through alternative splicing to inhibit mitochondrial apoptotic pathways and promote tumor formation (43). Additionally, the TOP2B gene encodes a DNA topoisomerase, which is involved in processes such as chromosome condensation, chromatid separation, and the relief of torsional stress that occurs during DNA transcription and replication (44). The ACLY gene can encode a central metabolic enzyme, which can catalyze the ATP-dependent conversion of citrate and coenzyme A (CoA) to oxaloacetate and acetyl-CoA. The acetyl-CoA product is crucial for the metabolism of fatty acids, the biosynthesis of cholesterol, and the acetylation and prenylation of proteins; thus, many cancer cells depend on the activity of ACLY for proliferation. It has been found to be overexpressed in several types of cancers with the "turning off" of ACLY being shown to lead to the cessation of cancer cell growth and division; ACLY has, therefore, become an important object of targeted therapies for cancer therapy research (45). EFTUD2 gene encodes a GTPase, a component of the spliceosome complex, which processes precursor mRNAs to produce mature mRNAs. SRSF1 can enhance epithelial cell metastasis, promote epithelial mesenchymal transition, and promote tumor formation (46). In addition, due to its special function being involved in alternative splicing, SRSF1 also promotes tumor formation and tumor proliferation by regulating alternative priming of other proto-oncogenes.
The analysis in this study showed that the low expression of SRSF1 could significantly improve the survival of breast cancer patients, which is consistent with the function observed in the previous study.
The changes in the expression of the hub gene affected by chickpea isoflavones can significantly improve the survival of patients. While the difference within 5 years was not particularly large, the survival extension observed beyond 5 years was significant. Therefore, we believe that chickpea isoflavones have certain preventive and therapeutic effects on MCF-7 type breast cancer, and thus have a role to play in long-term healthcare.
Conclusions
Through the integrated analysis of wet and dry experiments, we found that isoflavones in chickpeas may promote the apoptosis of MCF-7 cells by inhibiting the expression of HNRNP family genes mediated by lncRNAs. The up-regulation and down-regulation of hub genes in the PPI network triggered by chickpea isoflavones have positive effects on improving the survival time of breast cancer patients. These findings can provide a reference for developing chickpea isoflavones into new anti-breast cancer drugs.
Acknowledgments
Funding: The work was supported financially by Key Projects in the National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (2015BAD29B03-4), the Technology Program of Xinjiang Uygur Autonomous Region (201491160), the West Light Foundation of the Chinese Academy of Sciences (LHXZ-2014-01), and the National Key Research and Development Program of China Stem Cell and Translational Research (2017YFA0105101).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA-Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Burstein HJ, Temin S, Anderson H, et al. Adjuvant endocrine therapy for women with hormone receptor-positive breast cancer: american society of clinical oncology clinical practice guideline focused update. J Clin Oncol 2014;32:2255-69. [Crossref] [PubMed]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA-Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Deapen D, Liu L, Perkins C, et al. Rapidly rising breast cancer incidence rates among Asian-American women. Int J Cancer 2002;99:747-50. [Crossref] [PubMed]
- Gibson LJ, Héry C, Mitton N, et al. Risk factors for breast cancer among Filipino women in Manila. Int J Cancer 2010;126:515-21. [Crossref] [PubMed]
- Wu Z, Song L, Feng S, et al. Germination dramatically increases isoflavonoid content and diversity in chickpea (Cicer arietinum L.) seeds. J Agric Food Chem 2012;60:8606-15. [Crossref] [PubMed]
- HaiRong M. The estrogenic activity of isoflavones extracted from chickpea Cicer arietinum L sprouts in vitro. Phytother Res 2013;27:1237-42. [Crossref] [PubMed]
- Chen H, Ma HR, Gao YH, et al. Isoflavones extracted from chickpea Cicer arietinum L. sprouts induce mitochondria-dependent apoptosis in human breast cancer cells. Phytother Res 2015;29:210-9. [Crossref] [PubMed]
- Lv Q, Yang Y, Zhao Y, et al. Comparative Study on Separation and Purification of Isoflavones from the Seeds and Sprouts of Chickpea by HSCCC. J Liq Chromatogr Relat Technol 2009;32:2879-92. [Crossref] [PubMed]
- Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7:562-78. [Crossref] [PubMed]
- Gene OC, Mulder N. The Gene Ontology (GO) project in 2006. Nucleic Acids Res 2006;34:D322-6. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Spinozzi F, Pagliacci M C, Migliorati G, et al. The natural tyrosine kinase inhibitor genistein produces cell cycle arrest and apoptosis in Jurkat T-leukemia cells. Leuk Res 1994;18:431-9. [Crossref] [PubMed]
- Salti GI, Grewal S, Mehta RR, et al. Genistein induces apoptosis and topoisomerase II-mediated DNA breakage in colon cancer cells. Eur J Cancer 2000;36:796-802. [Crossref] [PubMed]
- Su SJ, Chow NH, Kung ML, et al. Effects of soy isoflavones on apoptosis induction and G2-M arrest in human hepatoma cells involvement of caspase-3 activation, Bcl-2 and Bcl-XL downregulation, and Cdc2 kinase activity. Nutr Cancer 2003;45:113-23. [Crossref] [PubMed]
- Mazzio E, Badisa R, Mack N, et al. High throughput screening of natural products for anti-mitotic effects in MDA-MB-231 human breast carcinoma cells. Phytother Res 2014;28:856-67. [Crossref] [PubMed]
- Yun SM, Jung JH, Jeong SJ, et al. Tanshinone IIA induces autophagic cell death via activation of AMPK and ERK and inhibition of mTOR and p70 S6K in KBM-5 leukemia cells. Phytother Res 2014;28:458-64. [Crossref] [PubMed]
- Chibbar RN, Ambigaipalan P, Hoover R. Molecular diversity in pulse seed starch and complex carbohydrates and its role in human nutrition and health. Cereal Chem 2010;87:342-52. [Crossref]
- Geervani P. Utilization of chickpea in India and scope for novel and alternative uses. Uses of Tropical Grain Legumes 1989;3:47-54.
- Roy F, Boye JI, Simpson BK. Bioactive proteins and peptides in pulse crops: Pea, chickpea and lentil. Food Res Int 2010;43:432-42. [Crossref]
- Wood JA, Grusak MA. Nutritional value of chickpea. Chickpea breeding and management 2007:101-42.
- FAOSTAT (2016). Available online: . (Accessed 15th December 2017)http://www.fao.org/faostat/en/#data/QC
- Visa N, Alzhanova-Ericsson AT, Sun X, et al. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell 1996;84:253-64. [Crossref] [PubMed]
- Eversole A, Maizels N. In Vitro Properties of the Conserved Mammalian Protein hnRNP D Suggest a Role in Telomere Maintenance. Mol Cell Biol 2000;20:5425. [Crossref] [PubMed]
- Kamma H, Fujimoto M, Fujiwara M, et al. Interaction of hnRNP A2/B1 isoforms with telomeric ssDNA and the in vitro function. Biochem Biophys Res Commun 2001;280:625-30. [Crossref] [PubMed]
- Ishikawa F, Matunis MJ, Dreyfuss G, et al. Nuclear proteins that bind the pre-mRNA 3'splice site sequence r (UUAG/G) and the human telomeric DNA sequence d (TTAGGG) n. Mol Cell Biol 1993;13:4301-10. [Crossref] [PubMed]
- LaBranche H, Dupuis S, Ben-David Y, et al. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nature Genet 1998;19:199. [Crossref] [PubMed]
- Moran-Jones K. hnRNP A2, a potential ssDNA/RNA molecular adapter at the telomere. Nucleic Acids Res 2005;33:486-96. [Crossref] [PubMed]
- Shi X, Ran L, Liu Y, et al. Knockdown of hnRNP A2/B1 inhibits cell proliferation, invasion and cell cycle triggering apoptosis in cervical cancer via PI3K/AKT signaling pathway. Oncol Rep 2018;39:939-50. [PubMed]
- Zhao S, Feng J, Wang Q, et al. HnRNP K plays a protective role in TNF-α-induced apoptosis in podocytes. Biosci Rep 2018;38:BSR20180288. [Crossref] [PubMed]
- Kędzierska H, Piekiełko-Witkowska A. Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett 2017;396:53-65. [Crossref] [PubMed]
- Tang Y, Geng Y, Luo J, et al. Downregulation of ubiquitin inhibits the proliferation and radioresistance of non-small cell lung cancer cells in vitro and in vivo. Sci Rep 2015;5:9476. [Crossref] [PubMed]
- Lord K A, Abdollahi A, Hoffman-Liebermann B, et al. Proto-oncogenes of the fos/jun family of transcription factors are positive regulators of myeloid differentiation. Mol Cell Biol 1993;13:841-51. [Crossref] [PubMed]
- Wen S, Wang X, Wang Y, et al. Nucleoside diphosphate kinase 2 confers acquired 5-fluorouracil resistance in colorectal cancer cells. Artif Cells Nanomed Biotechnol 2018;46:896-905. [Crossref] [PubMed]
- Wang Q, Li Q, Liu T, et al. Host Interaction Analysis of PA-N155 and PA-N182 in Chicken Cells Reveals an Essential Role of UBA52 for Replication of H5N1 Avian Influenza Virus. Front Microbiol 2018;9:936. [Crossref] [PubMed]
- De Moliner KL, Wolfson ML, Bizzozero NP, et al. Growth-associated protein-43 is degraded via the ubiquitin-proteasome system. J Neurosci Res 2005;79:652-60. [Crossref] [PubMed]
- Mazurek A, Park Y, Miething C, et al. Acquired Dependence of Acute Myeloid Leukemia on the DEAD-Box RNA Helicase DDX5. Cell Reports 2014;7:1887-99. [Crossref] [PubMed]
- Clark EL, Coulson A, Dalgliesh C, et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res 2008;68:7938-46. [Crossref] [PubMed]
- Mazurek A, Luo W, Krasnitz A, et al. DDX5 Regulates DNA Replication and Is Required for Cell Proliferation in a Subset of Breast Cancer Cells. Cancer Discov 2012;2:812-25. [Crossref] [PubMed]
- Lee T, Pelletier J. Dependence of p53-deficient cells on the DHX9 DExH-box helicase. Oncotarget 2017;8:30908-21. [PubMed]
- Guest ST, Kratche ZR, Bollig-Fischer A, et al. Two members of the TRiC chaperonin complex, CCT2 and TCP1 are essential for survival of breast cancer cells and are linked to driving oncogenes. Exp Cell Res 2015;332:223-35. [Crossref] [PubMed]
- Bassiouni R, Nemec K, Iketani A, et al. Chaperonin Containing-TCP-1 Protein Level in Breast Cancer Cells Predicts Therapeutic Application of a Cytotoxic Peptide. Clin Cancer Res 2016;22:4366. [Crossref] [PubMed]
- Zhan L. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell 2008;135:865-78. [Crossref] [PubMed]
- Lang AJ, Mirski SEL, Cummings HJ, et al. Structural organization of the human TOP2A and TOP2B genes. Gene 1998;221:255-66. [Crossref] [PubMed]
- Wei J, Leit S, Kuai J, et al. An allosteric mechanism for potent inhibition of human ATP-citrate lyase. Nature 2019;568:566. [Crossref] [PubMed]
- Karni R, de Stanchina E, Krainer AR, et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol 2007;14:185-93. [Crossref] [PubMed]


