Identification and analysis of dysregulated lncRNA and associated ceRNA in the pathogenesis of keloid
Introduction
Keloid is an excessive fibrosis disease caused by abnormal proliferation of collagen fibers following trauma. None of the many methods that have been used to treat keloid, including triamcinolone injection, radical therapy and surgery, have been found to be efficient. This stresses the need for further insight into the pathogenesis driving this disease. Recently, links have been identified between keloid and several SNPs and loci, including rs873549 at lq41, rs8032158 at 15p21.3,rs940187 and rsl511412 at 3q22.3 (1), and lq41 and 15p21.3 in the Chinese Han population (2). Furthermore, several signaling pathways were focused as plausible pathways involved in keloid formation, such as phosphatidylinositol-3-kinase (PI3K)/Akt (3), mitogen-activated protein kinase (MAPK) (4), and transforming growth factor-beta 1 (TGF-β1) (5). However, the genetic and epigenetic basis of keloid is yet to be detected, and a more meticulous understanding of its pathogenesis is required.
Non-coding RNAs (ncRNAs) including long non-coding RNA (lncRNA) and microRNA (miRNA) play important roles in the pathogenesis of various diseases (6). There is an increasing amount of evidence which highlights lncRNA’s key contribution to tumorigenesis through its modulation of oncogenic and tumor suppressor pathways in tumor biology (7), including keloid. For example, miRNA-21 may promote keloid genesis via the P13K/AKT signaling pathway (8). Additionally, nowadays, more and more researchers focus on a gene regulatory network based on ceRNA (competing endogenous RNA) and the interaction of messenger RNA (mRNA), lncRNA, circRNA, pseudogene, etc. Zhu et al. reported that via miR-200c, lncRNA-ATB can inhibit the expression of zinc finger protein 217 (ZNF217), thus regulating the secretion of TGF-β2 in keloid fibroblast. As such, the ZNF217/TGF-β2/miR-200c/lncRNA-ATB signaling pathway may be a potential biomarker for the pathogenesis of keloid (9).
In this study, we performed RNA-seq and miRNA-seq on keloid and normal skin samples. We systematically identified genome-wide dysregulated mRNA, lncRNA, and miRNA, constructed a ceRNA network and validated 2 pairs of ceRNAs in another sample size. Understanding these novel RNA interactions will lead to a better understanding of the molecular mechanism of keloid pathogenesis, and lay the foundation for further research in this area, especially for the development of novel treatment options.
Methods
Samples and clinical information
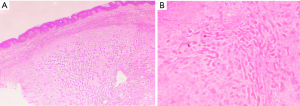
Thirteen patients with keloid tissue and 12 surrounding healthy tissue were recruited from Huashan Hospital at Fudan University. The diagnoses of the patients (mean ± SD age of 28.8±5.67 years) were confirmed by at least two dermatologists and assessed according to the Vancouver scar scale (VSS). The features of keloid tissues are depicted in Figure S1; briefly, these are abnormal accumulation of extracellular matrix (ECM) and excess proliferation of fibroblasts. All participants were required to sign informed consent documents. This research was conducted in accordance with the principles out lined in the Declaration of Helsinki.
RNA sequencing
Total RNA was isolated from tissue samples using TRIzol (Invitrogen, Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s included protocol. Total RNA was accessed by electrophoresis on a denaturing agarose gel and quantified by NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DL, USA). A TruSeq Small RNA Sample Preparation Kit (Illumina, San Diego, CA, USA) and a TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA) were used to generate the miRNA and RNA sequencing library. A Hiseq 4000 platform (Illumina, San Diego, CA, USA) was used to sequence the library preparations by 150 bp paired-end reads for RNA-seq and 50 bp of single-end reads for miRNA-seq.
Sequencing data analysis
We checked the quality of the data processing of raw reads using Fastqc (v0.10.1, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and, where necessary, used TrimGalore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) to trim low-quality bases (a quality of less than 10) and adaptors. We used HISAT2 (v2.0.4) to map clean reads to the reference genome for RNAseq, as well as using StringTie (v1.3.1) to assemble the mapped reads of each sample (10). Differential expression analysis of mRNA and lncRNA was conducted using the DESeq2 R package, and for miRNA-seq, Bowtie (v1.1.2) was used to map the clean reads to the hg19 reference genome. Known mature miRNA expression profiles were generated using miRDeep2. For normalization, we used the bioconductor package edgeR (11).
Functional enrichment analysis
To analyze the differential mRNAs and lncRNA-associated differential mRNAs, we used gene ontology (GO) analysis, and identified the significant pathway associated with differential mRNAs through Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Enrichment analysis of the GO and KEGG pathways was performed using the clusterProfiler R package.
ceRNA network analysis
The miRanda was used to predict miRNA/lncRNA interactions. A ceRNA network of mRNA/miRNA/lncRNA was generated by Cytoscape v3.0.
qRT-PCR validation
qRT-PCR was performed using a SYBR-Green PCR kit. The primers used for the qRT-PCR are detailed in Table S1.

Full table
Statistical analysis
Statistical analysis was performed using Statistical Programme for Social Sciences 20.0 (SPSS, Inc., Chicago, IL, USA), R Studio (R version 3.4.1). Differences in the qRT-PCR results were calculated by paired t-tests. The threshold used for statistically significant difference was P<0.05.
Results
mRNA, lncRNA and miRNA differential expression analysis (Figure 1)
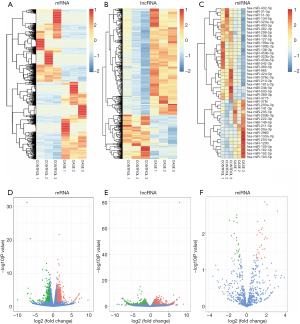
With a threshold of adjusted P value <0.05 and fold change >2 or <0.5, a total of 319 lncRNAs (68 downregulated and 251 upregulated) and 1,533 mRNAs (792 downregulated and 741 upregulated) between keloid samples and normal samples were detected in RNA-seq (Figure 1A,B,D,E). Thirteen downregulated miRNAs and 27 upregulated miRNAs were detected in miRNA-seq (Figure 1C,F). A hierarchical clustering heatmap and volcano plots were used to display the differentially expressed mRNAs, lncRNAs and miRNAs.
Functional enrichment analyses of keloid
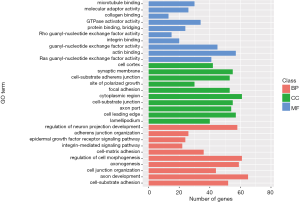
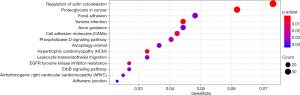
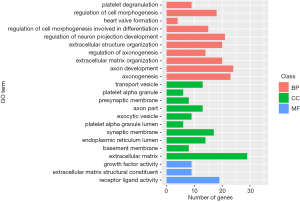
GO and KEGG enrichment analysis was performed on the 1,533 differentially expressed mRNAs to investigate their biological functions (Figures 2,3). The GO functional enrichment analysis explored BP, CC and MF. In BP analysis, microtubule binding, molecular adaptor activity and collagen binding were the three most enriched terms. In CC analysis, cell cortex, synaptic membrane and cell-substrate adherens junction were the three most enriched terms. In MF analysis, regulation of neuron projection development, adherens junction organization and the epidermal growth factor receptor signaling pathway were the three most enriched terms. The KEGG pathway enrichment analysis showed that differentially expressed mRNA were mainly enriched in 14 pathways, the top 3 of which were regulation of actin cytoskeleton, yersinia infection and proteoglycans in cancer (Table 1).

Full table
Co-expression analysis
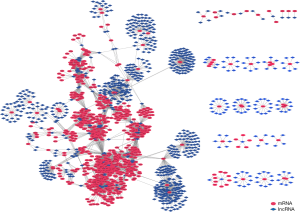
A correlation coefficient was calculated to identify the co-expression of lncRNAs with mRNAs, after filtering out the genes that were not significantly differentially expressed from all of the samples. Only the pairs of lncRNA and mRNA with adjusted P<0.05 were regarded as having a true correlation. Finally, 860 mRNAs and 835 lncRNAs with 6,728 correlations were retained to construct a co-expression network between lncRNA and mRNA (Figure 4). We also found that 207 differentially expressed lncRNAs interacted with 268 mRNAs in the co-expression network mainly enriched in GO term ECM and axonogenesis (Figure 5).
ceRNA network construction and analysis
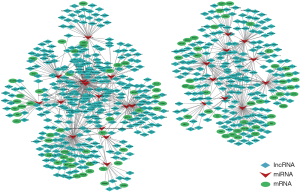
A ceRNA network was established by applying the theory that the binding of miRNA and target mRNA could lead to gene silencing, while ceRNA can play as a sponge to competitively bind with the same miRNA. Target mRNAs of miRNA were predicted by miRand, and we found 1,615 mRNAs and 49 miRNAs can be matched. Of these mRNAs, a negative correlation with miRNA expression levels was chosen. Target lncRNAs were selected using the method mentioned above. Based on the interaction of mRNA-miRNA, lncRNA-miRNA with shared miRNA, we constructed a general mRNA-miRNA-lncRNA network. Totally, we formed a ceRNA network consisting of 94 mRNAs, 509 lncRNA and 25 miRNAs (1,016 edges and 628 nodes, Figure 6). The intersection of the target mRNAs and differentially expressed mRNAs in keloid was identified.
qRT- PCR validation of transcriptome data of selected ceRNA networks
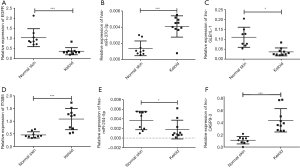
Among the 94 mRNAs in ceRNA, 5 genes—the epidermal growth factor receptor (EGFR), integrin-β5 (ITGB5), the S100 calcium binding protein A4 (S100A4), the molecule CD68, and galanin (GAL) (12-14)—have been reported to be associated with keloids. ITGB5 and EGFR were involved in the top enriched pathways of regulation of the actin cytoskeleton. Here, we selected EGFR and ITGB5 genes as candidate genes for validation of ceRNAs in keloid. For each mRNA, we found unique mRNA/miRNA interactions and multiple lncRNA/miRNA interactions. Since most of the lncRNAs in the selected ceRNA networks have low expression, lncRNAs were selected based on their high expression in RNA-seq data (reads >30), and with unique isoform. We chose 2 groups of ceRNA network: EGFR/miR-370-3p/lnc-GLB1L-1 and ITGB5/miR-204/lnc-CASP9-3 to perform qRT-PCR validation using samples from another 10 patients with keloid and 9 of nearby normal tissue. As shown in Figure 7, the expression of all of the selected ceRNAs was coincident with the result of RNA-seq, which confirmed the accuracy of sequencing. In the first ceRNA network: EGFR and lnc-GLB1L-1 was downregulated, miR-370-3p was upregulated in keloid tissue. In the second ceRNA network: ITGB5 and lnc-CASP9-3 was upregulated, miR-204 was downregulated in keloid tissue.
Discussion
Currently, large-scale cDNA sequencing and RNA sequencing already determines more than 10,000 lncRNAs in the mammalian genome, including humankind. The miRNAs and lncRNAs discussed make an important impact in the progression of multifarious diseases including keloid. For example, research conducted about lncRNA-ATB suggested a signaling axis composition of ZNF217/TGF-β2/miR-200c/lncRNA-ATB, and there has also been an association reported between keloids and CACNA1G-AS1, HOXA11-AS, LINC00312 and other lncRNAs (9,15). Nevertheless, the expression profiles of mRNA, lncRNA and miRNA remain elusive. In our study, we compared genome-wide expression profiles between the lesioned skin and normal skin of keloid patients. The differentially expressed mRNAs, lncRNAs and miRNAs were identified and analyzed to elucidate the pathogenesis of keloid.
It has been proven that genetics and epigenetics are clearly related to the pathogenesis of keloid. Until now, only one GWAS carried out on keloid analysis, and 4 SNPs (rs873549, rs1511412, rs940187 and rs8032158) were proven to be associated with keloid (1). We suspect that this relative lack of research into the genetic basis of keloid is at least part of the reason that we are unable to find significant associations between known SNPs and our sequencing data.
Our research found the most significant pathway to be the regulation of the actin cytoskeleton. A vast array of core eukaryotic phenotypes is governed by the actin cytoskeleton, including cell movement, endocytosis, vesicular trafficking, and cytokinesis (16). Consequently, cellular motility and the invasion of cancer also participated with the dynamic remodeling of actin filaments (17). This pathway can explain the invasive nature of keloids compared with hypertrophic scars. Regulation of the actin cytoskeleton pathway further relates to the adherens junction pathway, the MAPK signaling pathway and the focal adhesion pathway, which also participates in the generation and development of many human cancers. In this pathway, EGFR, ITGB5 were also reported to be associated with keloid. Based on the above analysis, we further selected 2 pairs of ceRNAs: EGFR/miR 370-3p/lnc GLB1L-1 and ITGB5/miR-204/lnc-CASP9-3 for relative expression qRT-PCR verification. The final quantitative results were consistent with the sequencing results, and successfully validate that lncRNA GLB1L-1 serving as a competitive endogenous RNA, downregulates EGFR expression via sponging miR-370-3p. Our results further validate that lncRNA CASP9-3 serving as a competitive endogenous RNA, upregulates ITGB5 expression via sponging miR-204.
EGFR belongs to the tyrosine kinase receptor family and has tyrosine kinase activity. When combined with the corresponding ligand EGF, it can activate the tyrosine protein kinase binding domain of the receptor, while further activating the downstream signal transduction pathway, which causes cell division and proliferation and increased DNA synthesis (18). EGFR is also a key gene in the mechanism of keloid development. Previous studies indicate that EGFR is less expressed and that there is a decrease of activation in keloid tissue (4). Besides, EGF-induced keloid tissue fibroblasts have less migration than normal tissue fibroblasts, and mitosis is reduced (19). This indicates that the low expression of EGFR promotes the development of keloid. MiR-370-3p is located on human chromosome 14 within the DLK1/DIO3 domain, which also considered to be a tumor-associated genomic region (20). Previous studies have shown that downregulation of miR-370-3p expression is relevant to ovarian cancer, cholangiocarcinoma, hepatocellular carcinoma, bladder cancer, oral squamous cell carcinoma, neuroblastoma cells and acute myeloid leukemia. In contrast, another study found that miR-370-3p is upregulated in gastric cancer, nephroblastoma, and prostate cancer, promoting the progression. In keloids, the biological and functional mechanisms involved in miR-370-3p are unknown. lncRNA GLB1L-1 is located on chromosome 2 (chr2: 220101439-220118807) and has a long-chain non-coding RNA of 17,369 bp in length. It has not yet been reported on. Its low expression in keloid tissue suggests that it may be associated with the occurrence of keloids. The mechanism is related, and further study into its specific regulatory mechanism is required.
ITGB5, which is a member of the integrin family, has been found to promote tumorigenesis in many cancers through its modulation of interaction between cells and the ECM. There is growing evidence which highlights the contribution of ITGB5 in cancer cell migration, invasion and angiogenesis (21,22). It also can be an attractive target for tumor therapy because of its correlation with oncogenes (23). The cooperation between ITGB5 and the endothelial growth factor receptor 2 (VEGF2) inhibits extrinsic apoptosis to enhance cell survival (24,25). ITGB5 also plays a pivotal role in epithelial-mesenchymal transition (EMT) through the transforming growth factor β (TGF-β) (26). Studies have shown that ITGB5 is significantly upregulated both in keloid and hypertrophic scar (27). MiR-204 is located on human chromosome 9. It specifically binds with target genes to regulate both the proliferation and differentiation of cells. Previous studies have shown that the expression of miR-204 was downregulated in gastric and thyroid cancer tissues (28). MiR-204 acts while the autophagy-related miRNAs participate in the growth of keloid fibroblast (29). lncRNA CASP9-3 is located on chromosome 1 (chr1: 15326680-15343876) and has a long-chain non-coding RNA of 3,105 bp in length. Furthermore, more experiments are required into lnc-CASP9-3 as a ceRNA of ITGB5 via miR-204.
In conclusion, our study analyzed the lncRNA, mRNA and miRNA expression profiles of keloid based on RNA-seq and miRNA-seq. The key lncRNA and miRNA which were identified in our study may serve as valuable diagnostic and therapeutic targets which could lay a foundation for further genetic research. Through co-expression analysis and ceRNA network construction, it was revealed that 2 groups of ceRNA networks: EGFR/miR-370-3p/lnc-GLB1L-1 and ITGB5/miR-204/lnc-CASP9-3 may participate in the pathogenesis of keloid.
Acknowledgments
Funding: This research is supported by the Natural Science Foundation of China (Grant No. 81402589) and the Key Laboratory of Dermatology of the Education Ministry of China, Anhui Medical University (Grant No. AY2017-1-008).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by Medical Ethics Committee of Huashan Hospital Affiliated to Fudan University (No. 2018-091). All participants were required to sign informed consent documents.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nakashima M, Chung S, Takahashi A, et al. A genome-wide association study identifies four susceptibility loci for keloid in the Japanese population. Nat Genet 2010;42:768-71. [Crossref] [PubMed]
- Zhu F, Wu B, Li P, et al. Association study confirmed susceptibility loci with keloid in the Chinese Han population. PLoS One 2013;8:e62377. [Crossref] [PubMed]
- An G, Liang S, Sheng C, et al. Upregulation of microRNA-205 suppresses vascular endothelial growth factor expression-mediated PI3K/Akt signaling transduction in human keloid fibroblasts. Exp Biol Med (Maywood) 2017;242:275-85. [Crossref] [PubMed]
- Satish L, Babu M, Tran KT, et al. Keloid fibroblast responsiveness to epidermal growth factor and activation of downstream intracellular signaling pathways. Wound Repair Regen 2004;12:183-92. [Crossref] [PubMed]
- Jagadeesan J, Bayat A. Transforming growth factor beta (TGFbeta) and keloid disease. Int J Surg 2007;5:278-85. [Crossref] [PubMed]
- Diamantopoulos MA, Tsiakanikas P, Scorilas A. Non-coding RNAs: the riddle of the transcriptome and their perspectives in cancer. Ann Transl Med 2018;6:241. [Crossref] [PubMed]
- Wang M, Sun X, Wang H, et al. Long non-coding RNAs in non-small cell lung cancer: functions and distinctions from other malignancies. Transl Cancer Res 2019;8:2636-53. [Crossref]
- Li Y, Zhang J, Lei Y, et al. MicroRNA-21 in Skin Fibrosis: Potential for Diagnosis and Treatment. Mol Diagn Ther 2017;21:633-42. [Crossref] [PubMed]
- Zhu HY, Bai WD, Li C, et al. Knockdown of lncRNA-ATB suppresses autocrine secretion of TGF-β2 by targeting ZNF217 via miR-200c in keloid fibroblasts. Sci Rep 2016;6:24728. [Crossref] [PubMed]
- Pertea M, Kim D, Pertea GM, et al. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 2016;11:1650-67. [Crossref] [PubMed]
- Wang L, Feng Z, Wang X, et al. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010;26:136-8. [Crossref] [PubMed]
- Hurwitz ZM, Ignotz R, Lalikos JF, et al. Accelerated porcine wound healing after treatment with α-gal nanoparticles. Plast Reconstr Surg 2012;129:242e-251e. [Crossref] [PubMed]
- Jiao H, Fan J, Cai J, et al. Analysis of Characteristics Similar to Autoimmune Disease in Keloid Patients. Aesthetic Plast Surg 2015;39:818-25. [Crossref] [PubMed]
- Xiaoxue W, Xi C, Zhibo X. Effects of Botulinum Toxin Type A on Expression of Genes in Keloid Fibroblasts. Aesthet Surg J 2014;34:154-9. [Crossref] [PubMed]
- Sun XJ, Wang Q, Guo B, et al. Identification of skin-related lncRNAs as potential biomarkers that involved in Wnt pathways in keloids. Oncotarget 2017;8:34236-44. [PubMed]
- Velle KB, Fritz-Laylin LK. Diversity and evolution of actin-dependent phenotypes. Curr Opin Genet Dev 2019;58-59:40-8. [Crossref] [PubMed]
- Ohishi T, Yoshida H, Katori M, et al. Tankyrase-Binding Protein TNKS1BP1 Regulates Actin Cytoskeleton Rearrangement and Cancer Cell Invasion. Cancer Res 2017;77:2328-38. [Crossref] [PubMed]
- Marmor MD, Skaria KB, Yarden Y. Signal transduction and oncogenesis by ErbB/HER receptors. Int J Radiat Oncol Biol Phys 2004;58:903-13. [Crossref] [PubMed]
- Kang SU, Kim YS, Kim YE, et al. Opposite effects of non-thermal plasma on cell migration and collagen production in keloid and normal fibroblasts. PLoS One 2017;12:e0187978. [Crossref] [PubMed]
- Kircher M, Bock C, Paulsen M. Structural conservation versus functional divergence of maternally expressed microRNAs in the Dlk1/Gtl2 imprinting region. BMC Genomics 2008;9:346. [Crossref] [PubMed]
- Su G, Hodnett M, Wu N, et al. Integrin alphavbeta5 regulates lung vascular permeability and pulmonary endothelial barrier function. Am J Respir Cell Mol Biol 2007;36:377-86. [Crossref] [PubMed]
- Li F, Liu Y, Kan X, et al. Elevated expression of integrin αv and β5 subunit in laryngeal squamous-cell carcinoma associated with lymphatic metastasis and angiogenesis. Pathol Res Pract 2013;209:105-9. [Crossref] [PubMed]
- Takada Y, Ye X, Simon S. The integrins. Genome Biol 2007;8:215. [Crossref] [PubMed]
- Hood JD, Frausto R, Kiosses WB, et al. Differential alphav integrin-mediated Ras-ERK signaling during two pathways of angiogenesis. J Cell Biol 2003;162:933-43. [Crossref] [PubMed]
- Tancioni I, Uryu S, Sulzmaier FJ, et al. FAK Inhibition disrupts a β5 integrin signaling axis controlling anchorage-independent ovarian carcinoma growth. Mol Cancer Ther 2014;13:2050-61. [Crossref] [PubMed]
- Bianchi A, Gervasi ME, Bakin A. Role of β5-integrin in epithelial-mesenchymal transition in response to TGF-β. Cell Cycle 2010;9:1647-59. [Crossref] [PubMed]
- Huang C, Nie F, Qin Z, et al. A Snapshot of Gene Expression Signatures Generated Using Microarray Datasets Associated With Excessive Scarring. Am J Dermatopathol 2013;35:64-73. [Crossref] [PubMed]
- Wu ZY, Wang SM, Chen ZH, et al. MiR-204 regulates HMGA2 expression and inhibits cell proliferation in human thyroid cancer. Cancer Biomark 2015;15:535-42. [Crossref] [PubMed]
- Gao J, Jiao H, Cao R, et al. In vitro study of effects and mechanism of rapamycin-induced autophagy in keloid fibroblasts. Zhonghua Zheng Xing Wai Ke Za Zhi 2016;32:208-14. [PubMed]