
Advances in laboratory assays for detecting human metapneumovirus
Introduction
Human metapneumovirus (HMPV) is one of the main viral etiological agents of acute respiratory tract infection (ARI), which is a major cause of morbidity and mortality, especially in southeast Asia (1). HMPV was first discovered by genetic analysis of nasopharyngeal aspirate samples taken from 28 hospitalized children in the Netherlands. These samples, taken from children with ARIs during the past 20 years, had symptoms ranging from mild respiratory problems to severe cough, bronchiolitis, and pneumonia. Some of them were hospitalized and needed mechanical ventilation (2). Since then, HMPV has been detected in approximately 6.39% of patients hospitalized with ARI (3). The prevalence of HMPV varied from 0% (4) to 36.4% (5), showing high heterogeneity in previous studies. HMPV is commonly found in children, particularly those less than 2 years old (6). HMPV infection has also affected immunocompromised patients such as elderly adults (7) and hematopoietic stem cell-transplant recipients (8). The HMPV outbreaks showed an overall fatality rate of 11% (9).
HMPV is an enveloped virus that is comprised of a negative-sense, single-stranded RNA genome. The HMPV genome harbors 8 genes that encode for 9 proteins. The order of these genes in the genome is N-P-M-F-M2-SH-G-L. The encoded proteins include the nucleoprotein (N protein), phosphoprotein (P protein), matrix protein (M protein), fusion glycoprotein (F protein), putative transcription factor (M2-1 protein), RNA synthesis regulatory factor (M2-2 protein), small hydrophobic glycoprotein (SH protein), attachment glycoprotein (G protein), and viral polymerase (L protein) (10,11). The F, M, and L genes as well as the N gene (the most conserved region), have been targeted to detect HMPV by reverse transcriptase-polymerase chain reaction (RT-PCR) (12,13). In the past few decades, several methods have been developed and established for detecting HMPV (6). Traditional diagnostic techniques (such as virus culture) and immunoassays have been used for pathogen detection. Recently, the advent of newly developed assays utilizing melting-curve analysis, and nucleic acid-amplification technologies has enabled clinical laboratories to detect HMPV rapidly and precisely. When adopting these assays, the comprehensive understanding of the principles, advantages, disadvantages, and precautions for data interpretation should be well-informed. Articles focusing on laboratory methods for HMPV detection have seldomly been published. Therefore, we reviewed the laboratory assays used for HMPV in the real-world laboratories based on published articles.
RT-PCR
RT-PCR is commonly used to detect HMPV (Table 1). This technique involves the reverse transcription of RNA into complementary DNA (cDNA) and the amplification of specific DNA targets using PCR. Simultaneous performance of RT-PCR and real-time PCR, which enables combined nucleic acid amplification and detection in a single step, is routinely used to detect viral RNA (14).
Full table
Nucleic acid extraction and purification are basic steps used for most RT-PCR systems. The sample concentration can be measured to improve the clinical sensitivity. These procedures are automated in numerous laboratories for high-quality and reproducible results. The QIAamp viral RNA Mini Kit from Qiagen (15-17), the MagNaPure Compact system from Roche (18,19), and the NucliSENS easyMAG platform from bioMérieux (20,21) are widely used for automated RNA extraction from respiratory swabs or aspirate specimens. Including control material is necessary to validate the extraction and purification steps. Primers and probes enabling amplification and detection of viral targets have been reported (15,22,23). Primers against conserved regions have been preferred for reliable amplification or for identifying sequence variants. However, these primers can cause decreased amplification efficiency when homologous primers incorporate into products. Several HMPV genes such as P, M, F, or N have served as targets for RT-PCR. Among them, the N gene is most conserved and has shown high sensitivity and reliability for all four genotypes; thus, it has been widely targeted in RT-PCR assays (12,13,24).
The PCR thermal cycling programs consist of three steps (denaturation, annealing, and elongation) and are slightly varied, according to previous reports for HMPV (15,23). Quantification can be performed by RT-PCR as either a one-step or a two-step reaction (25). One of the main differences between these two procedures is the number of used tubes. For one-step RT-PCR, most processes going from the reverse transcriptase reaction to PCR amplification are conducted in a single tube. However, cDNA synthesis and PCR amplification occur in separate tubes in two-step reactions. The advantage of using a one-step protocol is that it minimizes experimental variation. However, using RNA (which is prone to degradation) as the starting templates for one-step reactions makes it difficult to repeatedly assay the same samples over a period of time. In addition, one-step protocols are reported to be less sensitive than two-step protocols (26). Two-step reaction show high reproducibility and it is possible to perform several different PCR assays after diluting a single cDNA sample (27). Although two-step protocols are vulnerable to DNA contamination, they have been the preferred methods when using DNA-binding dyes such as SYBR Green. Primer dimers can be easily eliminated by manipulating the melting temperatures.
Gel-based detection (28), automated fluorescent capillary electrophoresis (29), or hybridization to target specific probes (30) have been used to detect the amplified products. Nested PCR was adopted in several laboratories because of its enhanced sensitivity and specificity (31,32). However, problems with amplicon contamination have occurred even in experienced laboratories (33). As a preferred method in clinical laboratories, real-time PCR assays utilize target-specific probes to detect the amplified products. Among various types of fluorescent probes used for real-time PCR, TaqMan and molecular beacon probes are most widely used. The fluorescent signals generated by TaqMan and molecular beacon probes depend on Förster resonance energy transfer (FRET)-based coupling of the dye molecule and a quencher moiety to the oligonucleotide substrate (34). During PCR amplification, TaqMan probes hybridize to the target sequences and cleave the fluorescent probe. Decoupling of the quencher molecule and the fluorescent probe increases the fluorescence intensity in direct proportion to the number of probes that are cleaved (35). TaqMan probes were frequently used to detect HMPV in previous studies (36-39). Molecular beacon probes also utilize FRET detection. However, they remain intact and rebind to a new target during each PCR cycle. When molecular beacon probes hybridize to a target, the fluorescent dye and the quencher are separated and emit fluorescent light upon excitation (40). Both TaqMan and molecular beacon probes are expensive and time-consuming to synthesize and require separate probes for each RNA target. HMPV detection was previously achieved using the NucliSENS EasyQ instrument (bioMérieux) and molecular beacon probes (3,41). The appropriateness of selecting the real-time amplification format for HMPV detection depends on the number of targets to be tested, laboratory throughput, level of expertise available, and acceptable hands-on or run time. The spectral overlap of fluorophores should be considered, especially for fluorescent real-time PCR assays. Checking the specificity of probe hybridization is recommended due to its usefulness and relative convenience.
Multiplex RT-PCR
In multiplex PCR, multiple primer sets are included within a single PCR tube to produce amplicons specific to different target sequences. The primer pairs should be optimally designed to function at the same annealing temperature. Different amplicons are detected using target-specific probes labeled with different fluorescent dyes (42). This technique enables detection of a broader range of respiratory viruses in a single test run. Additionally, information pertaining to coinfection can be obtained through multiplex PCR (35,43). A 30% to 50% increase in the detection of respiratory viruses was reported when using multiplex RT-PCR methods compared to conventional methods, such as viral culture or direct fluorescent antibodies (44). Commercial multiplex PCR kits, cleared by the Food and Drug Association (FDA), have become available, although they offer mixtures of pros and cons (45). Among them, the FilmArray system (BioFire Diagnostics, Inc.)—integrating nucleic acid extraction, nested PCR, and data analysis in a disposable pouch—enables HMPV detection. This system is recommended considering its short hands-on and run time, and reagent storage condition (room temperature). Limitations of the FilmArray system (such as the ability to only perform a single test per run) should also be considered when selecting an instrument(s) suitable for a clinical laboratory (45-47).
Introducing multiple primer and probe sets can enable the detection of multiple viral pathogens; however, the potential for reduced amplification efficiency exists. Separating the PCR amplification step from the hybridization and detection steps solve problems associated with diagnostic efficiency and enhances the capacity for detecting a broader array of pathogens. These separate hybridization techniques have become a forerunner of array-based methods. Microarrays comprise collections of microscopic DNA or RNA spots bound to a solid surface, with the potential of analyzing complex amplified PCR products. The microarray substrate can be fabricated from nylon, membrane, glass, silicon, or polystyrene microbeads, based on probe design and hybridization conditions. These microarray-based approaches have proven useful for detecting multiple respiratory viruses, including HMPV. High‐throughput gene analysis using a dynamic array of integrated fluidics chips on the BioMark platform (38,48), or the CLART PneumoVir platform from Genomica (49) have been utilized for HMPV detection. Furthermore, liquid-phase, bead-conjugated microarray methods have been applied with commercial Luminex systems for detecting amplified products. When adding new probes to solid-phase microarrays, it is necessary to reformat the procedures and print new arrays, although these steps are not required for suspension microarrays. Therefore, liquid-phase microarray techniques exhibit flexibility in terms of assay design and format, rapid hybridization kinetics, and lower cost (50,51). The Luminex xTAG RVP fast assay is a multiplexed microsphere-based suspension microarray platform that enables the analysis and reporting of 12 respiratory viruses from nasopharyngeal swabs. Several investigators have adopted the commercially available Luminex system for HMPV detection (52-54). The Luminex system showed higher positivity (47.5%) than real-time RT-PCR (42.5%) and immunofluorescence assays (2.7%) (55). However, as previously mentioned, the FilmArray system was reported to have higher sensitivity than the Luminex xTAG RVP fast assay and shorter a run time (1 hour) and hands-on time (2 minutes) (45,52). Additional enzymatic amplification steps for viral RNA prior to hybridization are required for microarray-based methods. Moreover, detection requires labeling of multiple probes or the incorporation of fluorescent dye- or biotin-conjugated nucleotides into cDNA. In addition, the sensitivities of most conventional microarrays have depended on the efficiency of target amplification and probe hybridization. The multiple steps required for these microarrays make them complex, expensive, labor-intensive, and vulnerable to contamination. These array systems are also prone to false-negative results because of the gene mutations, PCR inhibitors, and degradation of RNA products. Further investigations are required to optimize the design of multiple, virus-specific primers and assay systems.
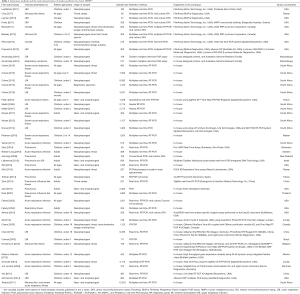
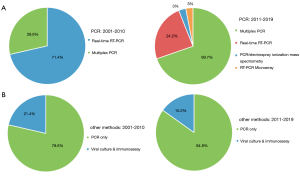
The detection methods used and their main characteristics observed in previous HMPV studies are presented in Table 1. The included studies were mainly based on four HMPV meta-analyses recently published in 3 years [2017–2019] (45,56-58). Forty-seven studies are included to reflect methods used in clinical laboratories. The countries/areas included in this study were Belgium (59), Botswana (60), Brazil (61), Canada (22), China (23,39,62), Denmark (38), Israel (37), Jordan (19,63), Kenya (36), Malawi (64), Mexico (48), Mozambique (65,66), New Zealand (67), South Africa (18,31,32,68-76), Sweden (77), Taiwan (17,78), Thailand (15), Turkey (49), and USA (3,7,16,20,21,52-54,79-83). The most frequently used method was multiplex PCR (57.4%), followed by real-time RT-PCR (38.3%) (Figure 1). Commercially available products from diverse companies, such as the One-Step RT-PCR Kit (Applied Biosystems or Qiagen), TaqMan real-time PCR kits, NucliSENS EasyQ instrument (bioMérieux), and Pro hMPV Real Time assay (Gen-Probe) were used to detect HMPV. The differences in the assays used to detect HMPV according to previous publications are illustrated in Figure 2. Multiplex PCR (69.7%) became the most commonly used method in 2011–2019, in contrast to 2001–2009. The advent of multiplex PCR for detecting multiple pathogens (including coinfecting viruses) in a single run influenced these changes in user preferences.
Viral culture
The isolation of viruses from culture is considered a gold standard for diagnosing infection (52,64). After inoculating a permissive cell line with an infectious sample, such as a nasopharyngeal swab or aspirate, the specimen is incubated for 7 to 10 days to observe the development of a cytopathic effect (CPE). CPE refers to structural changes in cells caused by viral invasion. Established cell lines, such as Madin-Darby canine kidney cells, A549 cells, mink lung epithelial cell lines, human lung diploid fibroblast cells (MRC-5 and WI-38), HeLa cells, rhesus monkey kidney cells (LLC-MK2), and buffalo green monkey kidney cells are utilized for isolating diverse viruses, including influenza virus and respiratory syncytial virus (52,64,84). Regarding HMPV, various cell lines, such as the Vero (85), HEp-2, Hep G2 (86), 293 (87), and LLC-MK2 (2,7,52) cell lines have been used to isolate the virus. Among them, a human Chang conjunctiva cell line (clone 1-5C4) and a feline kidney CRFK cell line were suggested as the most suitable cells for HMPV isolation, based on a previous study of 19 different cell lines used to grow HMPV (88). HMPV culture in LLC-MK2 cell lines has been frequently performed in clinical laboratories, based on previous reports (7,52). The tolerance of LLC-MK2 cells to trypsin make it an ideal cell line for HMPV cultivation (85).
Traditional viral culture has been generally replaced by shell vial culture. This technique involves the inoculation of samples onto a cell monolayer in shell vials, followed by centrifugation and further incubation. After 24 to 48 hours, the HMPV antigens are measured with virus-specific antibodies. Shell vial culture enables rapid detection of slow-growing viruses, including HMPV. Moreover, this method is relatively straightforward and more sensitive than traditional viral culture, due to the centrifugation steps (89). In addition, a modified shell vial culture method has been introduced. This technique, utilizing R-Mix or R-Mix Too cells from Diagnostic Hybrids (a mixture of mink lung cells and human adenocarcinoma cells), has shown higher sensitivity than the shell vial culture method with a run time of 1.4 days (80,84).
HMPV grows so slowly that it shows late CPEs varying from cell rounding and detachment from the culture matrix to the formation of small syncytia (i.e., large cytoplasmic masses that contain many nuclei). Therefore, immunoassays such as direct fluorescent assays (DFAs) and enzyme-linked immunosorbent assays (ELISAs) are frequently utilized along with cell culture to detect HMPV antigens (85).
Florescent immunoassay
Among the various florescent immunoassays, DFA testing directly determines the presence of specific antigens with fluorescently labeled antibodies and has been widely employed to detect viruses in clinical laboratories. This method involves the direct staining of respiratory epithelial cells derived from nasopharyngeal swabs or aspirates with fluorescently labeled, virus-specific antibodies (82,90). DFA methods coupled with viral culture or shell vial culture are also widely used for enhanced sensitivity and specificity (80). Data interpretation using immunofluorescence microscopy is necessary. Determination of the correct absorption wavelength is required to excite the fluorophore tag bound to the antibody and detect the released fluorescence, where positive cells are indicated by the presence of the targeted virus. DFAs have been used for several decades due to their simplicity and short turnaround time. A modified cytospin-enhanced DFA has also been used to enhance the sensitivity in terms of virus detection. Cytospinning decreases inadequate smears and improves cell morphology, resulting in enhanced performance. Data from a previous study showed that 85.4% (41/48) of samples were positive by cytospin-enhanced DFA, compared to TaqMan RT-PCR (91). Among commercially available DFA kits approved by the FDA, the D3 DFA test (Diagnostic Hybrids) has been widely used (44,80). This kit enables the identification of HMPV in less than 25 minutes after sample receipt and shows high sensitivity (95.2%) and specificity (100%) compared to RT-PCR (92). When compared to shell vial culture, DFA showed comparable results (93.3%) based on the number of positive samples determined by RT-PCR (93). In another study, comparing viral culture with DFA showed a 92% sensitivity and a 90% specificity for HMPV (80). However, no recent studies have involved HMPV detection without molecular techniques because of the shorter turnaround times and higher sensitivity of molecular tools.
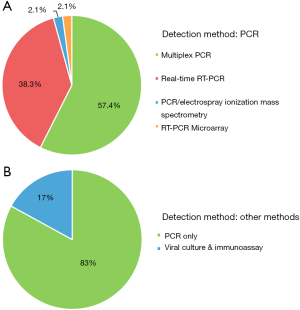
The assays used and their main characteristics in studies involving viral culture and fluorescent immunoassays are shown in Table 1. Among 47 included studies, 8 studies (17.0%) involved viral culture and fluorescent immunoassays, whereas the remaining 39 studies (83.0%) employed only PCR-based methods for HMPV detection (Figure 1). LLC-MK2 cells (Diagnostic Hybrids) have been frequently used for HMPV culture in several clinical laboratories (7,52). The usefulness of trypsin-tolerant LLC-MK2 cells for HMPV cultivation has influenced these trends (85). In terms of DFAs, the FDA-approved D3 DFA kit from Diagnostic Hybrids have been widely used (44,80). The short turnaround time (less than 25 minutes) and high performance compared to RT-PCR make it commonly utilized. Changes over time in the use of culture and fluorescent immunoassays for detecting HMPV are presented in Figure 2. The combined proportions of culture-based and fluorescent immunoassays decreased from 21.4% in 2001–2010 to 15.2% in 2011–2019. Meanwhile, PCR-based methods showed an increasing trend (78.6% in 2001–2010 versus 84.8% in 2011–2019). These changes might have occurred because PCR methods such as RT-PCR and multiplex PCR demonstrated better diagnostic performances (45). In a previous study, the sensitivity and specificity of viral culture were reported to be 68% and 99%, respectively, as compared to real-time RT-PCR detection of HMPV (94). In addition, the shorter run- and hands-on times, and more reliable results than traditional methods make PCR techniques preferable tools in many clinical laboratories (45).
Other methods
Among traditional methods for detecting HMPV, serological assays including virus-neutralization assays (95-97) and enzyme immunoassays (7) were once the main techniques. Virus-neutralization assays are used to detect virus-specific antibodies induced by viral infection. Virus-specific antibodies are produced by the immune system to neutralize viruses. The titer can be measured and the highest serum dilution at which virus infection is blocked is considered as the virus-neutralization titer. When performing virus-neutralization assays, the hazards due to the use of infectious viruses should considered. Several studies have been performed to develop better neutralization assays for HMPV detection (95-97). Plaque-reduction, virus-neutralization assays showed faster turnaround times and better sensitivities than conventional methods requiring around 7 days for completion (95). Micro-neutralization assays for measuring antibody titers (96) and neutralization assay based on recombinant HMPV expressing Renilla luciferase (97) also enabled efficient HMPV detection.
In terms of enzyme immunoassays, enzymes are conjugated to secondary (detection) antibodies, which bind to the primary viral antigen-antibody complex. When the appropriate substrate is added and incubated, the enzyme catalyzes the production of a colored end-product, which can be visualized and quantified. The most frequently used enzymes are alkaline phosphatase, horseradish peroxidase, and β-galactosidase (98). It is necessary to distinguish specific HMPV antigens from nonspecific complexes, for most commercially available systems. These systems are known as ELISA or solid-phase immunosorbent assays. Separation is feasible through binding of the antigen or capturing the antibody on a solid material, such as a microtiter plate or paper strip. Lysates from representative HMPV strains (7) or the carboxy-terminal domain of the N protein of HMPV (99) can be used for antigen preparation. Although ELISA-based tests have been developed with rapid turnaround times (100) and better diagnostic performance than traditional methods, PCR-based assays offering higher sensitivity have become preferred techniques in clinical laboratories.
In terms of modified PCR-based methods, the PCR-electrospray ionization mass spectrometry (ESI-MS) approach provided by the PLEX-ID system (Abbott Laboratories) was applied to detect HMPV. After RT-PCR, automated post-PCR desalting, ESI-MS signal acquisition, spectral analysis, and data reporting can be performed on a biosensor platform (Ibis Biosciences, USA). Virus identification was based on the MS data and base compositions of the PCR amplicons when compared to those in the molecular database established by the PLEX-ID manufacturer (17,101). Accurate identification based on both MS data and base compositions of PCR amplicons are an advantage of the ESI-MS technique. However, the process involves a longer turnaround time from sample to result (within 6 to 8 hours) and is more laborious than currently available multiplex RT-PCR systems (45). Regarding advanced multiplex RT-PCR methods, microarrays using nanoparticles coupled with silver staining have been adopted and served as tools for detecting viruses. This nanoparticle-based microarray offers better sensitivity than fluorescent dyes commonly utilized in most microarrays. When combining with next-generation sequencing (NGS) technology, this assay can be suitable for screening and identifying a broader range of viruses with shorter run- and hands-on times, due to the simplified procedures (102). This approach has been investigated mainly for influenza viruses and further studies with HMPV are necessary.
NGS has provided a quantum leap in the field of molecular diagnostics. NGS offers significant improvements in sequencing speed and throughput due to automated streamline workflow and reduces the cost of sequencing. Several manufacturer-specific platforms such as the Roche 454 system and the Illumina MiSeq and HiSeq systems (which use different sequencing strategies, reagents, and bioinformatics software) have been developed. Among them, Illumina’s NGS platforms (employing a sequencing-by-synthesis approach) have been used to analyze small non-coding RNAs of HMPV (103) and HMPV viral metagenomics (104).
Conclusions and future perspectives
In conclusion, we reviewed applied laboratory methodologies for detecting HMPV, one of the major causes of morbidity and mortality derived from ARI. In clinical laboratories, the main methods used for HMPV detection are PCR-based assays including conventional RT-PCR, multiplex RT-PCR, and microarray-based approaches. Viral culture such as shell vial assays and fluorescent immunoassays (combined with or without culture) were once the major methods for detecting HMPV. Comparison of the assays using the wide range of available data showed that PCR-based methods have become the more appreciated methods. Although viral culture and immunoassays have remained in use as reference methods due to their high specificity, PCR-based methods offer shorter run- and hands-on times, higher sensitivity, a broader range of available viral targets, a lower risk of exposure to virus, and economic feasibility, which could have influenced the changes in use. When using these assays, it is important to have a comprehensive understanding of the associated principles, advantages, disadvantages, and precautions for data interpretation.
Sophisticated techniques for detecting HMPV including nanotechnology and biosensors, with micro-sized instruments, ultra-sensitive and specific detectors, faster turnaround times, and lowered costs will be developed in the future. Furthermore, rapid and accurate diagnosis based on the advent of newly developed methods for differentiating subtypes and emerging viral variants resistant to currently used therapies will greatly benefit patients with HMPV infections. In addition, the genetic profiling of putative virulence factors and antiviral resistance markers using molecular tools such as NGS might improve patient prognosis and increase the accuracy in predicting responses to therapeutic intervention. Analytical and clinical validation should be conducted before using each new technique in clinical laboratories.
Acknowledgments
The authors would like to thank Hyun Jung Kim (Korea University College of Medicine) and the Biomedical Research Institute (Pusan National University Hospital) for providing technical support and statistical consultation.
Funding: This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science, ICT & Future Planning) (NRF-2017R1C1B2004597). The funder was not involved in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Zhi-De Hu, Bing Gu) for the series “Advances in laboratory tests for infectious diseases” published in Annals of Translational Medicine. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2019.12.42). The series “Advances in Laboratory Tests for Infectious Diseases” was commissioned by the editorial office without any funding or sponsorship. SJ reports grants from National Research Foundation of Korea, during the conduct of the study. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. The other authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Williams BG, Gouws E, Boschi-Pinto C, et al. Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect Dis 2002;2:25-32. [Crossref] [PubMed]
- van den Hoogen BG, de Jong JC, Groen J, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 2001;7:719-24. [Crossref] [PubMed]
- Akhras N, Weinberg JB, Newton D. Human metapneumovirus and respiratory syncytial virus: subtle differences but comparable severity. Infect Dis Rep 2010;2:e12. [Crossref] [PubMed]
- Ouédraogo S, Traore B, Nene Bi ZA, et al. Viral etiology of respiratory tract infections in children at the pediatric hospital in Ouagadougou (Burkina Faso). PLoS One 2014;9:e110435. [Crossref] [PubMed]
- Smuts H, Workman L, Zar HJ. Role of human metapneumovirus, human coronavirus NL63 and human bocavirus in infants and young children with acute wheezing. J Med Virol 2008;80:906-12. [Crossref] [PubMed]
- Panda S, Mohakud NK, Pena L, et al. Human metapneumovirus: review of an important respiratory pathogen. Int J Infect Dis 2014;25:45-52. [Crossref] [PubMed]
- Falsey AR, Erdman D, Anderson LJ, et al. Human metapneumovirus infections in young and elderly adults. J Infect Dis 2003;187:785-90. [Crossref] [PubMed]
- Dokos C, Masjosthusmann K, Rellensmann G, et al. Fatal human metapneumovirus infection following allogeneic hematopoietic stem cell transplantation. Transpl Infect Dis 2013;15:E97-101. [Crossref] [PubMed]
- Centers for Disease Control and Prevention (CDC). Outbreaks of human metapneumovirus in two skilled nursing facilities -West Virginia and Idaho, 2011-2012. MMWR Morb Mortal Wkly Rep 2013;62:909-13. [PubMed]
- Biacchesi S, Murphy BR, Collins PL, et al. Frequent frameshift and point mutations in the SH gene of human metapneumovirus passaged in vitro. J Virol 2007;81:6057-67. [Crossref] [PubMed]
- van den Hoogen BG, Bestebroer TM, Osterhaus AD, et al. Analysis of the genomic sequence of a human metapneumovirus. Virology 2002;295:119-32. [Crossref] [PubMed]
- Lefebvre A, Manoha C, Bour JB, et al. Human metapneumovirus in patients hospitalized with acute respiratory infections: A meta-analysis. J Clin Virol 2016;81:68-77. [Crossref] [PubMed]
- Maertzdorf J, Wang CK, Brown JB, et al. Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J Clin Microbiol 2004;42:981-6. [Crossref] [PubMed]
- Freeman WM, Walker SJ, Vrana KE. Quantitative RT-PCR: pitfalls and potential. Biotechniques 1999;26:112-22, 24-5.
- Dare RK, Fry AM, Chittaganpitch M, et al. Human coronavirus infections in rural Thailand: a comprehensive study using real-time reverse-transcription polymerase chain reaction assays. J Infect Dis 2007;196:1321-8. [Crossref] [PubMed]
- Self WH, Williams DJ, Zhu Y, et al. Respiratory Viral Detection in Children and Adults: Comparing Asymptomatic Controls and Patients With Community-Acquired Pneumonia. J Infect Dis 2016;213:584-91. [Crossref] [PubMed]
- Shih HI, Wang HC, Su IJ, et al. Viral Respiratory Tract Infections in Adult Patients Attending Outpatient and Emergency Departments, Taiwan, 2012-2013: A PCR/Electrospray Ionization Mass Spectrometry Study. Medicine (Baltimore) 2015;94:e1545. [Crossref] [PubMed]
- Moyes J, Cohen C, Pretorius M, et al. Epidemiology of respiratory syncytial virus-associated acute lower respiratory tract infection hospitalizations among HIV-infected and HIV-uninfected South African children, 2010-2011. J Infect Dis 2013;208 Suppl 3:S217-26. [Crossref] [PubMed]
- Schuster JE, Khuri-Bulos N, Faouri S, et al. Human Metapneumovirus Infection in Jordanian Children: Epidemiology and Risk Factors for Severe Disease. Pediatr Infect Dis J 2015;34:1335-41. [Crossref] [PubMed]
- Jain S, Self WH, Wunderink RG, et al. Community-Acquired Pneumonia Requiring Hospitalization among U.S. Adults. N Engl J Med 2015;373:415-27. [Crossref] [PubMed]
- Zimmerman RK, Rinaldo CR, Nowalk MP, et al. Influenza and other respiratory virus infections in outpatients with medically attended acute respiratory infection during the 2011-12 influenza season. Influenza Other Respir Viruses 2014;8:397-405. [Crossref] [PubMed]
- Chano F, Rousseau C, Laferriere C, et al. Epidemiological survey of human metapneumovirus infection in a large pediatric tertiary care center. J Clin Microbiol 2005;43:5520-5. [Crossref] [PubMed]
- Yan XL, Li YN, Tang YJ, et al. Clinical characteristics and viral load of respiratory syncytial virus and human metapneumovirus in children hospitaled for acute lower respiratory tract infection. J Med Virol 2017;89:589-97. [Crossref] [PubMed]
- Dare R, Sanghavi S, Bullotta A, et al. Diagnosis of human metapneumovirus infection in immunosuppressed lung transplant recipients and children evaluated for pertussis. J Clin Microbiol 2007;45:548-52. [Crossref] [PubMed]
- Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques 2005;39:75-85. [Crossref] [PubMed]
- Battaglia M, Pedrazzoli P, Palermo B, et al. Epithelial tumour cell detection and the unsolved problems of nested RT-PCR: a new sensitive one step method without false positive results. Bone Marrow Transplant 1998;22:693-8. [Crossref] [PubMed]
- Vandesompele J, De Paepe A, Speleman F. Elimination of primer-dimer artifacts and genomic coamplification using a two-step SYBR green I real-time RT-PCR. Anal Biochem 2002;303:95-8. [Crossref] [PubMed]
- Coyle PV, Ong GM, O'Neill HJ, et al. A touchdown nucleic acid amplification protocol as an alternative to culture backup for immunofluorescence in the routine diagnosis of acute viral respiratory tract infections. BMC Microbiol 2004;4:41. [Crossref] [PubMed]
- Erdman DD, Weinberg GA, Edwards KM, et al. GeneScan reverse transcription-PCR assay for detection of six common respiratory viruses in young children hospitalized with acute respiratory illness. J Clin Microbiol 2003;41:4298-303. [Crossref] [PubMed]
- Coiras MT, Lopez-Huertas MR, Lopez-Campos G, et al. Oligonucleotide array for simultaneous detection of respiratory viruses using a reverse-line blot hybridization assay. J Med Virol 2005;76:256-64. [Crossref] [PubMed]
- Madhi SA, Ludewick H, Kuwanda L, et al. Pneumococcal coinfection with human metapneumovirus. J Infect Dis 2006;193:1236-43. [Crossref] [PubMed]
- Madhi SA, Ludewick H, Kuwanda L, et al. Seasonality, incidence, and repeat human metapneumovirus lower respiratory tract infections in an area with a high prevalence of human immunodeficiency virus type-1 infection. Pediatr Infect Dis J 2007;26:693-9. [Crossref] [PubMed]
- Apfalter P, Reischl U, Hammerschlag MR. In-house nucleic acid amplification assays in research: how much quality control is needed before one can rely upon the results? J Clin Microbiol 2005;43:5835-41. [Crossref] [PubMed]
- Holden MJ, Wang L. Quantitative Real-Time PCR: Fluorescent Probe Options and Issues. Standardization and Quality Assurance in Fluorescence Measurements II. Springer Series on Fluorescence. Springer, 2008.
- Li H, McCormac MA, Estes RW, et al. Simultaneous detection and high-throughput identification of a panel of RNA viruses causing respiratory tract infections. J Clin Microbiol 2007;45:2105-9. [Crossref] [PubMed]
- Feikin DR, Njenga MK, Bigogo G, et al. Etiology and Incidence of viral and bacterial acute respiratory illness among older children and adults in rural western Kenya, 2007-2010. PLoS One 2012;7:e43656. [Crossref] [PubMed]
- Lieberman D, Shimoni A, Shemer-Avni Y, et al. Respiratory viruses in adults with community-acquired pneumonia. Chest 2010;138:811-6. [Crossref] [PubMed]
- von Linstow ML, Larsen HH, Eugen-Olsen J, et al. Human metapneumovirus and respiratory syncytial virus in hospitalized danish children with acute respiratory tract infection. Scand J Infect Dis 2004;36:578-84. [Crossref] [PubMed]
- Zhan Y, Yang Z, Chen R, et al. Respiratory virus is a real pathogen in immunocompetent community-acquired pneumonia: comparing to influenza like illness and volunteer controls. BMC Pulm Med 2014;14:144. [Crossref] [PubMed]
- Cecchinato M, Lupini C, Munoz Pogoreltseva OS, et al. Development of a real-time RT-PCR assay for the simultaneous identification, quantitation and differentiation of avian metapneumovirus subtypes A and B. Avian Pathol 2013;42:283-9. [Crossref] [PubMed]
- Ginocchio CC, Manji R, Lotlikar M, et al. Clinical evaluation of NucliSENS magnetic extraction and NucliSENS analyte-specific reagents for real-time detection of human metapneumovirus in pediatric respiratory specimens. J Clin Microbiol 2008;46:1274-80. [Crossref] [PubMed]
- Briese T, Palacios G, Kokoris M, et al. Diagnostic system for rapid and sensitive differential detection of pathogens. Emerg Infect Dis 2005;11:310-3. [Crossref] [PubMed]
- Paranhos-Baccalà G, Komurian-Pradel F, Richard N, et al. Mixed respiratory virus infections. J Clin Virol 2008;43:407-10. [Crossref] [PubMed]
- Babady NE. The FilmArray® respiratory panel: an automated, broadly multiplexed molecular test for the rapid and accurate detection of respiratory pathogens. Expert Rev Mol Diagn 2013;13:779-88. [Crossref] [PubMed]
- Huang HS, Tsai CL, Chang J, et al. Multiplex PCR system for the rapid diagnosis of respiratory virus infection: systematic review and meta-analysis. Clin Microbiol Infect 2018;24:1055-63. [Crossref] [PubMed]
- Caliendo AM. Multiplex PCR and emerging technologies for the detection of respiratory pathogens. Clin Infect Dis 2011;52 Suppl 4:S326-30. [Crossref] [PubMed]
- Tao Y, Tang M, Luo L, et al. Identification of etiologic agents and clinical characteristics for patients suspected of having pertussis in a large Children’s Hospital in China. Ann Transl Med 2019;7:443. [Crossref] [PubMed]
- Moreno-Valencia Y, Hernandez-Hernandez VA, Romero-Espinoza JAI, et al. Detection and characterization of respiratory viruses causing acute respiratory illness and asthma exacerbation in children during three different seasons (2011-2014) in Mexico City. Influenza Other Respir Viruses 2015;9:287-92. [Crossref] [PubMed]
- Tokman HB, Aslan M, Ortakoylu G, et al. Microorganisms in respiratory tract of patients diagnosed with atypical pneumonia: results of a research based on the use of reverse transcription polymerase chain reaction (RT-PCR) DNA microarray method and enzyme-linked immunosorbent assay. Clin Lab 2014;60:1027-34. [Crossref] [PubMed]
- Dunbar SA. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin Chim Acta 2006;363:71-82. [Crossref] [PubMed]
- Mezger A, Kuhnemund M, Nilsson M, et al. Highly specific DNA detection employing ligation on suspension bead array readout. N Biotechnol 2015;32:504-10. [Crossref] [PubMed]
- Babady NE, Mead P, Stiles J, et al. Comparison of the Luminex xTAG RVP Fast assay and the Idaho Technology FilmArray RP assay for detection of respiratory viruses in pediatric patients at a cancer hospital. J Clin Microbiol 2012;50:2282-8. [Crossref] [PubMed]
- Popowitch EB, O'Neill SS, Miller MB. Comparison of the Biofire FilmArray RP, Genmark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP fast multiplex assays for detection of respiratory viruses. J Clin Microbiol 2013;51:1528-33. [Crossref] [PubMed]
- Rand KH, Rampersaud H, Houck HJ. Comparison of two multiplex methods for detection of respiratory viruses: FilmArray RP and xTAG RVP. J Clin Microbiol 2011;49:2449-53. [Crossref] [PubMed]
- Luchsinger V, Prades Y, Ruiz M, et al. Comparison of Luminex xTAG(R) RVP fast assay and real time RT-PCR for the detection of respiratory viruses in adults with community-acquired pneumonia. J Med Virol 2016;88:1173-9. [Crossref] [PubMed]
- Kenmoe S, Bigna JJ, Fatawou Modiyingi A, et al. Case fatality rate and viral aetiologies of acute respiratory tract infections in HIV positive and negative people in Africa: The VARIAFRICA-HIV systematic review and meta-analysis. J Clin Virol 2019;117:96-102. [Crossref] [PubMed]
- Ma X, Conrad T, Alchikh M, et al. Can we distinguish respiratory viral infections based on clinical features? A prospective pediatric cohort compared to systematic literature review. Rev Med Virol 2018;28:e1997. [Crossref] [PubMed]
- Shi T, Arnott A, Semogas I, et al. The Etiological Role of Common Respiratory Viruses in Acute Respiratory Infections in Older Adults: A Systematic Review and Meta-analysis. J Infect Dis 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Hombrouck A, Sabbe M, Van Casteren V, et al. Viral aetiology of influenza-like illness in Belgium during the influenza A(H1N1)2009 pandemic. Eur J Clin Microbiol Infect Dis 2012;31:999-1007. [Crossref] [PubMed]
- Zash RM, Shapiro RL, Leidner J, et al. The aetiology of diarrhoea, pneumonia and respiratory colonization of HIV-exposed infants randomized to breast- or formula-feeding. Paediatr Int Child Health 2016;36:189-97. [Crossref] [PubMed]
- Cuevas LE, Nasser AM, Dove W, et al. Human metapneumovirus and respiratory syncytial virus, Brazil. Emerg Infect Dis 2003;9:1626-8. [Crossref] [PubMed]
- Chen X, Zhang ZY, Zhao Y, et al. Acute lower respiratory tract infections by human metapneumovirus in children in Southwest China: a 2-year study. Pediatr Pulmonol 2010;45:824-31. [Crossref] [PubMed]
- Ali SA, Williams JV, Chen Q, et al. Human metapneumovirus in hospitalized children in Amman, Jordan. J Med Virol 2010;82:1012-6. [Crossref] [PubMed]
- Peterson I, Bar-Zeev N, Kennedy N, et al. Respiratory Virus-Associated Severe Acute Respiratory Illness and Viral Clustering in Malawian Children in a Setting With a High Prevalence of HIV Infection, Malaria, and Malnutrition. J Infect Dis 2016;214:1700-11. [Crossref] [PubMed]
- Annamalay AA, Abbott S, Sikazwe C, et al. Respiratory viruses in young South African children with acute lower respiratory infections and interactions with HIV. J Clin Virol 2016;81:58-63. [Crossref] [PubMed]
- Annamalay AA, Lanaspa M, Khoo SK, et al. Rhinovirus species and clinical features in children hospitalised with pneumonia from Mozambique. Trop Med Int Health 2016;21:1171-80. [Crossref] [PubMed]
- Jennings LC, Anderson TP, Beynon KA, et al. Incidence and characteristics of viral community-acquired pneumonia in adults. Thorax 2008;63:42-8. [Crossref] [PubMed]
- Cohen AL, Sahr PK, Treurnicht F, et al. Parainfluenza Virus Infection Among Human Immunodeficiency Virus (HIV)-Infected and HIV-Uninfected Children and Adults Hospitalized for Severe Acute Respiratory Illness in South Africa, 2009-2014. Open Forum Infect Dis 2015;2:ofv139. [Crossref] [PubMed]
- Cohen C, Moyes J, Tempia S, et al. Mortality amongst patients with influenza-associated severe acute respiratory illness, South Africa, 2009-2013. PLoS One 2015;10:e0118884. [Crossref] [PubMed]
- Cohen C, Moyes J, Tempia S, et al. Epidemiology of Acute Lower Respiratory Tract Infection in HIV-Exposed Uninfected Infants. Pediatrics 2016. [Crossref] [PubMed]
- Cohen C, Walaza S, Moyes J, et al. Epidemiology of viral-associated acute lower respiratory tract infection among children <5 years of age in a high HIV prevalence setting, South Africa, 2009-2012. Pediatr Infect Dis J 2015;34:66-72. [Crossref] [PubMed]
- Cohen C, Walaza S, Moyes J, et al. Epidemiology of severe acute respiratory illness (SARI) among adults and children aged >/=5 years in a high HIV-prevalence setting, 2009-2012. PLoS One 2015;10:e0117716. [Crossref] [PubMed]
- Moyes J, Walaza S, Pretorius M, et al. Respiratory syncytial virus in adults with severe acute respiratory illness in a high HIV prevalence setting. J Infect 2017;75:346-55. [Crossref] [PubMed]
- Nunes MC, Kuschner Z, Rabede Z, et al. Clinical epidemiology of bocavirus, rhinovirus, two polyomaviruses and four coronaviruses in HIV-infected and HIV-uninfected South African children. PLoS One 2014;9:e86448. [Crossref] [PubMed]
- Tempia S, Walaza S, Moyes J, et al. Attributable Fraction of Influenza Virus Detection to Mild and Severe Respiratory Illnesses in HIV-Infected and HIV-Uninfected Patients, South Africa, 2012-2016. Emerg Infect Dis 2017;23:1124-32. [Crossref] [PubMed]
- Venter M, Lassauniere R, Kresfelder TL, et al. Contribution of common and recently described respiratory viruses to annual hospitalizations in children in South Africa. J Med Virol 2011;83:1458-68. [Crossref] [PubMed]
- Brittain-Long R, Westin J, Olofsson S, et al. Prospective evaluation of a novel multiplex real-time PCR assay for detection of fifteen respiratory pathogens-duration of symptoms significantly affects detection rate. J Clin Virol 2010;47:263-7. [Crossref] [PubMed]
- Chiu SC, Lin YC, Wang HC, et al. Surveillance of upper respiratory infections using a new multiplex PCR assay compared to conventional methods during the influenza season in Taiwan. Int J Infect Dis 2017;61:97-102. [Crossref] [PubMed]
- Falsey AR, Criddle MC, Walsh EE. Detection of respiratory syncytial virus and human metapneumovirus by reverse transcription polymerase chain reaction in adults with and without respiratory illness. J Clin Virol 2006;35:46-50. [Crossref] [PubMed]
- Layman CP, Gordon SM, Elegino-Steffens DU, et al. Rapid multiplex PCR assay to identify respiratory viral pathogens: moving forward diagnosing the common cold. Hawaii J Med Public Health 2013;72:24-6. [PubMed]
- Loeffelholz MJ, Pong DL, Pyles RB, et al. Comparison of the FilmArray Respiratory Panel and Prodesse real-time PCR assays for detection of respiratory pathogens. J Clin Microbiol 2011;49:4083-8. [Crossref] [PubMed]
- Pierce VM, Elkan M, Leet M, et al. Comparison of the Idaho Technology FilmArray system to real-time PCR for detection of respiratory pathogens in children. J Clin Microbiol 2012;50:364-71. [Crossref] [PubMed]
- Poritz MA, Blaschke AJ, Byington CL, et al. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One 2011;6:e26047. [Crossref] [PubMed]
- Vemula SV, Zhao J, Liu J, et al. Current Approaches for Diagnosis of Influenza Virus Infections in Humans. Viruses 2016;8:96. [Crossref] [PubMed]
- Tollefson SJ, Cox RG, Williams JV. Studies of culture conditions and environmental stability of human metapneumovirus. Virus Res 2010;151:54-9. [Crossref] [PubMed]
- Schildgen V, Lusebrink J, Ditt V, et al. Human HepG2 cells support respiratory syncytial virus and human metapneumovirus replication. J Virol Methods 2010;163:74-81. [Crossref] [PubMed]
- Boivin G, Abed Y, Pelletier G, et al. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J Infect Dis 2002;186:1330-4. [Crossref] [PubMed]
- Isaeva EI, Kozulina IS, Podcherniaeva R, et al. Reproduction of the metapneumovirus in different cell lines. Vopr Virusol 2012;57:19-23. [PubMed]
- Jayakeerthi RS, Potula RV, Srinivasan S, et al. Shell Vial Culture assay for the rapid diagnosis of Japanese encephalitis, West Nile and Dengue-2 viral encephalitis. Virol J 2006;3:2. [Crossref] [PubMed]
- Ge X, Han Z, Chen H, et al. Characterization of acute respiratory infections among 340 infants in Wuxi, Jiangsu Province. Ann Transl Med 2015;3:264. [PubMed]
- Landry ML, Cohen S, Ferguson D. Prospective study of human metapneumovirus detection in clinical samples by use of light diagnostics direct immunofluorescence reagent and real-time PCR. J Clin Microbiol 2008;46:1098-100. [Crossref] [PubMed]
- Vinh DC, Newby D, Charest H, et al. Evaluation of a commercial direct fluorescent-antibody assay for human metapneumovirus in respiratory specimens. J Clin Microbiol 2008;46:1840-1. [Crossref] [PubMed]
- Jun KR, Woo YD, Sung H, et al. Detection of human metapneumovirus by direct antigen test and shell vial cultures using immunofluorescent antibody staining. J Virol Methods 2008;152:109-11. [Crossref] [PubMed]
- Matsuzaki Y, Mizuta K, Takashita E, et al. Comparison of virus isolation using the Vero E6 cell line with real-time RT-PCR assay for the detection of human metapneumovirus. BMC Infect Dis 2010;10:170. [Crossref] [PubMed]
- de Graaf M, Herfst S, Schrauwen EJ, et al. An improved plaque reduction virus neutralization assay for human metapneumovirus. J Virol Methods 2007;143:169-74. [Crossref] [PubMed]
- Falsey AR, Formica MA, Walsh EE. Microneutralization assay for the measurement of neutralizing antibodies to human metapneumovirus. J Clin Virol 2009;46:314-7. [Crossref] [PubMed]
- Zhou M, Kitagawa Y, Yamaguchi M, et al. Expeditious neutralization assay for human metapneumovirus based on a recombinant virus expressing Renilla luciferase. J Clin Virol 2013;56:31-6. [Crossref] [PubMed]
- Koivunen ME, Krogsrud RL. Principles of Immunochemical Techniques Used in Clinical Laboratories. Laboratory Medicine 2006;37:490-7. [Crossref]
- Zhang Y, Pohl J, Brooks WA, et al. Serologic cross-reactions between nucleocapsid proteins of human respiratory syncytial virus and human metapneumovirus. J Clin Microbiol 2015;53:1609-15. [Crossref] [PubMed]
- Ebihara T, Endo R, Ma X, et al. Detection of human metapneumovirus antigens in nasopharyngeal secretions by an immunofluorescent-antibody test. J Clin Microbiol 2005;43:1138-41. [Crossref] [PubMed]
- Chen KF, Rothman RE, Ramachandran P, et al. Rapid identification viruses from nasal pharyngeal aspirates in acute viral respiratory infections by RT-PCR and electrospray ionization mass spectrometry. J Virol Methods 2011;173:60-6. [Crossref] [PubMed]
- Zhao J, Ragupathy V, Liu J, et al. Nanomicroarray and multiplex next-generation sequencing for simultaneous identification and characterization of influenza viruses. Emerg Infect Dis 2015;21:400-8. [Crossref] [PubMed]
- Baños-Lara MDR, Zabaleta J, Garai J, et al. Comparative analysis of miRNA profile in human dendritic cells infected with respiratory syncytial virus and human metapneumovirus. BMC Res Notes 2018;11:432. [Crossref] [PubMed]
- Zhang D, Lou X, Yan H, et al. Metagenomic analysis of viral nucleic acid extraction methods in respiratory clinical samples. BMC Genomics 2018;19:773. [Crossref] [PubMed]


