
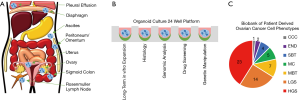
Tumor modeling maintains diverse pathology in vitro
Ovarian cancers present with clinical, molecular, and genetic heterogeneity. The diversity of ovarian tumors can be illustrated by the assortment of histological subtypes, sites of origin of the primary and metastatic lesions, and cellular makeup found within each patient. Known as the most lethal gynecological malignancies, they display disparate intra- and inter-tumoral heterogeneity, thereby making the pathology difficult to model. Standard 2D in vitro culture—using ovarian cancer cell lines fail to mimic the genetic diversity found within patient tumors. Ideally new cell lines would be established for each ovarian cancer patient, allowing for a personalized and individual approach to treatment. Unfortunately creating new cell lines is a challenging process, one that is both time-consuming and expensive. Moreover, when a new cell line is finally established, a strong in vitro selection can extensively deteriorate the purity of a tumor sample. Extended culture has been linked to both a loss of tumor molecular characteristic and copy number variance such that the resulting line becomes a tenuous representation of the original tissue sample. Kopper et al. (1) address these limitations by demonstrating a 3D method for growing heterogeneous organoids, modeling a diverse set of ovarian patient tumors. They show that the in vitro cancer models, produced from their organoid culturing system, maintain the diverse molecular and genetic traits of the original tumor sites. The authors establish an ovarian cancer biobank and use it to investigate promising leads, and attempt to ameliorate clinical challenges facing the field today.
Cytoreductive surgery followed by adjuvant carboplatin/paclitaxel chemotherapy has remained the standard-of-care treatment for ovarian cancer, however even with this aggressive treatment most patients develop recurrent disease within 12 to 18 months of their initial diagnosis. Although targeted therapies including angiogenesis and Poly (ADP-ribose) Polymerase (PARP) inhibitors have garnered much excitement, development of rapid chemoresistance necessitates novel therapies and physiologically relevant models to screen them. Conventional 2D models using established cell lines, have proven monumental in the advancement of cancer biology, physiology, biochemistry and cancer development. However, these cultures often lead to clonal expansion, genomic instability, and lack of cell heterogeneity. Additionally, due to significant rates of copy number mutations, and a lack of somatic mutations to stratify ovarian cancer patients, personalized drugs and their combinations are critically needed. Using a one size fits all method results in loss of representation of the tumor subtype’s specific characteristics, which often expand on hallmark mutations, morphology, signaling network, and tumorigenicity. Moreover traditional in vitro culture is not physiologically relevant as it lacks a tumor microenvironment, which contributes significantly to metastasis, chemoresistance and reoccurrence (2). 3D spheroids are an especially relevant ovarian cancer model, as they represent the physiological microtissues in the peritoneal ascites that circulate in the omentum of HGSOC patients, and are deemed critical to metastasis cascade. In vitro 3D spheroid models are a high throughput; non-adherent platform that can be generated from a number of sources. Spheroids are typically derived after the mechanical or enzymatic digestion of tumor tissue samples into single cell suspensions to form spheroids, which can be composed of multiple cell types. Organoids are a cluster of cells that proliferate and self-organize to form a 3D structure. Often these cell types originate from a single progenitor cell source. The organization of an organoid is very similar to its native tissue (3). Organoids provide a high throughput platform, essential to personalized medicine as they capture and maintain the heterogeneity of the tumor from which they are derived.
In vivo animal models including, patient derived xenografts (PDX) and genetically modified mouse models (GEMM) are an indispensable preclinical tool in ovarian cancer research. These models provide tumor cells with a partial physiologically relevant microenvironment, a preserved tumor architecture, and the ability to perform serial engraftment. Unfortunately, xenograft models are both time and resource intensive and force tumor cells to undergo mouse specific tumor evolution (4).
To address these shortcomings, the authors develop a robust organoid-culturing platform. Pertinent in its ability to recapitulate tumors in vitro which span the breadth of ovarian cancer subtypes. Organoid lines from a variety of patients were established using a 24-well plate culturing system, precoated with basement membrane extract (BME) and optimized using a custom ovarian cancer organoid medium. The authors took tissue from 7 different histological tumor sites, as well as tissue samples from the fimbria of the fallopian tube (FT) and the ovarian surface epithelium (OSE), culminating in 56 separate organoid lines from 32 patients that crossed the entirety of the disease’s developmental range.
The three main clinical challenges facing ovarian gynecological oncologists today include lack of early detection, tumor recurrence, and acquired chemoresistance. The author’s organoid platform enables long-term (validated up to passage 32) in vitro expansion of ovarian cancer subtypes. The tumor models produced are an accurate representation of original tissue samples for further use in analysis and manipulation. The author’s organoids maintain tumor histological characteristics, ovarian cancer hallmarks, recurrent mutations, tumor heterogeneity, biomarker expression, and genomic stability over extended passaging. Using 3D organoid methods to establish new representative tumor models, there is hope that these clinical challenges may yet be addressed.
As seen in Figure 1, a thorough analysis was performed on the organoid culture to prove the validity of their methodology and ensure the maintenance of heterogeneity over extended passaging. Tumor morphology and biomarker expression was maintained and the organoid lines faithfully modeled the original ovarian cancer at the genomic level shown through metaphase spread analysis, whole genome sequencing, and DNA methylation analysis. Organoids preserved intrapatient heterogeneity showing copy number changes within a single patient depended on the primary metastatic site of origin. Tumor heterogeneity was also assessed through novel single-cell DNA sequencing confirming consistency with the developed organoid line. Organoid gene expression profiles clustered with their respective primary sample throughout passaging and organoid lines derived from the same patient were found to be more related than inter-patient comparisons.
In addition to genomic data from each organoid, the researchers also profiled RNA transcripts identifying tumor lesions that were transcriptionally similar to one-another in each patient. Second round sequencing of later passages from multiple patients provided a biological link between non-malignant mucinous borderline tumors (MBT) and malignant mucinous (MC) samples supporting the progression hypothesis toward invasive carcinomas. The authors also found a tie between OSE organoids that clustered with ovarian cancer organoids instead of other OSE and FT groups; possibly identifying a malignancy that had remained undiagnosed by routine clinical examination, further emphasizing the clinical significance of this culture method.
Drug treatment investigations were performed on new organoid lines allowing for resistance and chemosensitivity to be monitored in vitro. Response rates of the organoids faithfully replicated matched tumor samples, while recurrent lines actually mirrored resistance observed in the clinic, demonstrating the practical use of this method for patient specific treatments. Proof of concept experiments shows the capability for genome editing using CRISPR-Cas9. Normal FT organoids were transduced with plasmids encoding for TP53 gene and then clonally expanded. An accompanying change in morphology was observed, including a transition from cystic to a denser organoid structure with an increase in cell shedding around the organoid lumen.
In this organoid model, the organoids were grown from isolated tumor cells; however, cancer does not grow in isolation and the effects of the various other supporting cells within the tumor microenvironment should be considered (5). Within the peritoneal fluid there are detached tumor cells, host cells including innate and adaptive immune cells, and activated mesothelial cells, all which can have cell-cell interactions with tumor cells (6). Additionally, the interactions of mesenchymal stem cells (MSC) and ovarian cancer cells have been widely studied, and the presence of MSCs can lead to increased cancer cell migration, metastasis, and resistance to chemotherapy (7). Furthermore, during passaging, the organoids were mechanically sheared through pipet tips. Multiple studies on the effect of shear stress on ovarian cancer have shown shear stress increased stress fiber formation and EMT biomarkers which can influence chemoresistance of cells, and thus should not be ignored (2). The choice of whether to use Wnt-condition medium or medium without Wnt conditioning was based on the organoid growth rate after two to three passages, and did not take into account the whether the components were found naturally in the body.
While the study was able to establish 56 organoid lines from thirty-two patients, only one patient’s cell lines were used when determining whether the organoids captured tumor heterogeneity. Also, when testing the xenotransplantation of the organoids only seven organoid lines were tested, and all three lines transplanted orthotopically were HGS. Furthermore, researchers concluded that the organoids recapitulated in vivo drug sensitivity in vitro after only studying one cell line in an in vivo mouse model. A greater variety of organoid cell lines analyzed for tumor heterogeneity, transplantation, and in vivo drug sensitivity would provide a more robust data set from which to draw conclusions.
Overall, Kopper et al. presented a reliable experimental model to address important clinical challenges, such as early detection, recurrence, and development of chemoresistance. They demonstrated a remarkable success rate, forming organoid lines from all subtypes of ovarian cancer as well as the FT and OSE. Using their model, organoid lines could be expanded, maintained, and stored in a biobank, facilitating downstream analysis of each sample. Importantly, they were able to show that their organoids maintained histological and genomic similarity to raw patient samples, even after passaging in vitro. Specifically, hallmarks of ovarian cancer were maintained including copy number variations, recurrent mutations, tumor heterogeneity. Furthermore, they showed that clustering of gene expression data grouped samples together according to ovarian cancer subtype, indicating maintenance of distinct subtype characteristics. The authors then demonstrated that these organoid models could be manipulated genetically, used in physiologically representative drug screens in vitro and in vivo, and used to identify malignant cells in samples from high-risk individuals.
Taken together, this model has enormous potential to address clinical challenges in ovarian cancer research. While not entirely novel due to the previous development of organoid model systems, this work stands out in the field as a result of the quality and quantity of the analysis and the long-term culture success that was achieved with a wide spectrum of patient samples. This is especially important given that patient samples are hard to obtain and often limited in their utility because they are so scarce. Using this model in ovarian cancer research could open the door to development of a comprehensive biobank that would accelerate all aspects of ovarian cancer research. Specifically, this model has potential applications in personalized medicine, early detection, and research on tumor recurrence and chemoresistance development, which are critical to improving clinical outcomes for ovarian cancer patients.
Now that Kopper et al. have thoroughly characterized their model system, further evaluation of its downstream applications are needed to maximize the overall impact on clinical outcomes and ovarian cancer research. For example, the authors demonstrated the value of their model as a tool for research on ovarian cancer development by identifying malignancy in an OSE sample (OSE(P)7) obtained from a prophylactic bilateral salpingo-oophorectomy (pBSO) and showing that they could use CRISPR-Cas9 genome editing on these normal samples to potentially induce tumor initiating mutations. Following through on this work with more samples from pBSOs will help to validate the use of this model in ovarian cancer development and early detection research. Based on the clustering of OSE(P)7 near a clinically diagnosed LGS-2 sample, further validation may help reveal a genetic signature of pre-malignant lesions that could facilitate development of preventative or early stage treatments.
Similarly, the authors demonstrated the utility of their model for drug screening and xenotransplantation applications, however further testing of their organoids with drug treatments equivalent to those used in their corresponding patient will be required to truly know the potential of the model for personalized medicine applications. To this end, it is critical to track patient outcomes and treatment methods whenever possible to allow for evaluation of the clinical relevance of drug screens. It will also be important to examine the response of organoids derived from multiple different locations in the tumor to treatment to evaluate the effect that intratumor heterogeneity may be having on recurrence and therapy resistance.
In addition to continually developing their biobank, it would be valuable for the authors to test if any of their organoid lines can be cultured under different conditions to evaluate how their culture system may be influencing their findings. Non-adherent environments would be especially important to test as ovarian cancer spheroids can often be found floating in the malignant ascites of ovarian cancer patients and can contribute to metastasis and recurrence (8). Moreover, while the presented organoids enrich for tumor cells, it would be valuable to characterize the initial cellular makeup of each tumor to evaluate how stromal, immune, and vascular cells may be influencing patient drug responses. This could also help to explain potential differences in drug response when comparing their organoids to patient outcomes. While we propose many future steps to maximize the utility of this organoid model system, the number of potential directions is indicative of the model’s potential in ovarian cancer research. In summary, this model is well developed and we believe that continued biobanking, analysis of downstream applications, and development of similar models for other cancers, could improve clinical outcomes for oncology patients around the world.
Acknowledgments
Funding: This work is supported primarily by DOD OCRP Early Career Investigator Award W81XWH-13-1-0134 (G Mehta), DOD Pilot award W81XWH-16-1-0426 (G Mehta), DOD Investigator Initiated award W81XWH-17-OCRP-IIRA (G Mehta), Rivkin Center for Ovarian Cancer (G Mehta) and Michigan Ovarian Cancer Alliance (G Mehta). The authors have no conflicts of interest to declare.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Kopper O, de Witte CJ, Lõhmussaar K, et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat Med 2019;25:838-49. [Crossref] [PubMed]
- Novak C, Horst E, Mehta G. Review: Mechanotransduction in ovarian cancer: Shearing into the unknown. APL Bioeng 2018;2:031701. [Crossref] [PubMed]
- Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009;459:262-5. [Crossref] [PubMed]
- Ben-David U, Ha G, Tseng YY, et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet 2017;49:1567-75. [Crossref] [PubMed]
- Bregenzer ME, Horst EN, Mehta P, et al. Integrated cancer tissue engineering models for precision medicine. PLoS One 2019;14:e0216564. [Crossref] [PubMed]
- Pogge von Strandmann E, Reinartz S, Wager U, et al. Tumor-Host Cell Interactions in Ovarian Cancer: Pathways to Therapy Failure. Trends Cancer 2017;3:137-48. [Crossref] [PubMed]
- Lis R, Touboul C, Halabi NM, et al. Mesenchymal cell interaction with ovarian cancer cells induces a background dependent pro-metastatic transcriptomic profile. J Transl Med 2014;12:59. [Crossref] [PubMed]
- Kim S, Kim B, Song YS. Ascites modulates cancer cell behavior, contributing to tumor heterogeneity in ovarian cancer. Cancer Sci 2016;107:1173-8. [Crossref] [PubMed]


