DNA methylation-based prognostic biomarkers of acute myeloid leukemia patients
Introduction
This study aims to analyze acute myeloid leukemia (AML); a group of genetically heterogeneous malignant clonal diseases that can block normal myeloid differentiation (1), as well as AML which is also the most common type of adult leukemia with the poorest prognosis.
Only in the United States of 2019, there have been 10,920 deaths, and 21,450 new cases of AML diagnosed (2). At present, induction therapy for AML is based primarily on cytotoxic drugs that enable complete remission (CR) in 70% of adult patients (3). However, the likelihood of the recurrence rate in AML patients is still high, especially in older patients with prognosis of having high-risk factors. Yet, the long-term cure rate was lower in the absence of allogeneic hematopoietic stem cell transplantation (Allo-HCT) in adult AML patients with first complete remission (CR1) (4).
With the emergence of various molecular targeted therapies for AML in the last two years, there are now several new drugs and other clinical trials currently underway; however, there is not enough data to know which newly approved drugs should be chosen or in which order or combination they should be used in (2). New early diagnostic biomarkers and therapeutic targets are therefore urgently needed to improve the diagnosis and risk management of AML in order to reduce the mortality rate of AML.
Epigenetic modifications include a class of heritable non-genetic changes in gene expression, usually including DNA methylation, histone modifications, and chromatin remodeling (5). In healthy hematopoietic stem cells, epigenetic processes play a key role in cell differentiation and hematopoiesis (6). Among them, DNA methylation affects the function of key genes. It is closely related to tumors by silencing tumor suppressor genes and activating oncogenes by high/low methylation (7). Abnormal DNA methylation is considered a hallmark of AML and is considered a powerful epigenetic marker in early diagnosis, prognosis prediction, and treatment decision making (8).
Abnormal gene expression is closely related to tumor prognosis. Studies have shown that NPM1 (9), FLT3 (10,11), C-KIT (12), AML1-ETO (13), RUNX1 (3,14), TP53 (3,15), CBFB/MYH11 (13,16), TET2 (17), DNMT3A (18), JAK-STAT (19), and CXCR4 (20,21), HOXA family (22), NAT10 (23) gene are associated with prognosis of AML. A few studies have shown altered DNA methylation in cancer, but the roles of key differentially methylated genes (DMGs) and differentially expressed genes (DEGs) in AML remain unclear.
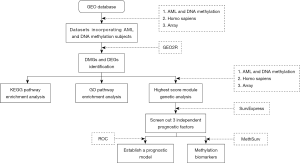
In this study, we performed a comprehensive analysis of DNA methylation and gene expression to identify key epigenetic genes in AML. The methylation genes and differential genes of AML patients and normal individuals were downloaded from the GEO database. After data preprocessing, we identified differential genes between tumors and normal samples and performed KEGG and GO analyses on these genes. Protein-protein interaction (PPI) network construction and module analysis were then performed, and the highest-scoring modules were screened. SurvExpress software and analyzed the genes to be assigned the highest-scoring module with a P value <0.05 were selected to perform survival analysis and risk assessment in the cancer dataset. Finally, MethSurv analyzed the three genes screened by SurvExpress software to explore methylation biomarkers associated with AML survival.
Methods
Microarray data
We extracted gene expression (GSE114868) and methylation (GSE63409) profiling data from the Gene Expression Omnibus (GEO) database at the National Center for Biotechnology Information.
The AML-associated dataset GSE63409 submitted by Jung N based on the GPL13534 platform was obtained from the GEO database and included 15 AML samples and 5 normal samples. The AML-associated dataset GSE114868 submitted by Huang H based on the GPL17586 platform was obtained from the GEO database and included 194 AML samples and 20 normal samples (Figure 1).
Identification of DEGs
GEO provides users with a useful tool called GEO2R that can be used to analyze microarray data. GRO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) were used to analyze gene expression in GSE114868 and GSE63409. Gene expression (GSE114868) and methylation (GSE63409) data set genes were considered statistically significant as |t| ≥2.0 and adj P<0.01. Then, we identified hypomethylated/upregulated genes via overlapping the hypomethylated and overexpression gene lists and identified hypermethylated/downregulated genes via overlapping the hypermethylated and low-expression gene lists. Finally, Then, we use the Venn diagram network tool to draw the Venn diagram. (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Functional enrichment analyses for module genes
The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database resource for understanding high-level functions and utilities of the biological system from molecular-level information. The Gene Ontology (GO) could be used to perform enrichment analysis. We used DAVID (https://david.ncifcrf.gov/) to make KEGG pathway analysis and GO enrichment analysis, including Biological Process, Cellular Component, and Molecular Function for the hypomethylated/upregulated genes and hypermethylated/downregulated genes.
PPI network construction and module analysis
PPI analysis is used to search core genes and gene modules related to carcinogenesis. In this study, the PPI network analysis of the hypomethylated/upregulated genes and hypermethylated/downregulated genes were performed using the search tool for the Retrieval of Interacting Genes (STRING) database. The interaction score was set at 0.7. Then, MCODE plugin in Cytoscape is used to find clusters with the degree cut-off, haircut on, k-core, node score cut-off, max depth set as 10, 0.2, 2, 0.2 and 100 in PPI network and the module with the highest score would be picked out to make survival analyses.
Survival analysis and Cox regression, ROC curve
SurvExpress is a comprehensive gene expression database and web-based tool that uses biomarker gene lists as input to provide survival analysis and risk assessment in cancer data sets (24). We selected genes in the highest-scoring module of the hypermethylated low expression and low methylation high expression overlapping genes, respectively. The univariate Cox model was used to calculate the association between gene expression levels and patient overall survival (OS). When the P value was <0.05, its modular gene was used as an independent prognostic factor for patient survival. AML samples are divided into two groups: (I) high-risk group; (II) low-risk group. Survival analysis was performed by Kaplan-Meier survival plots, log-rank P value, and hazard ratio (HR, 95% confidence interval). By comparing the sensitivity and specificity of risk-based survival predictions, the accuracy of prognostic performance was assessed using ROC curves of patients over time, 10, 30, 50, and 70 months, respectively.
DNA methylation data in MethSurv
MethSurv is used to explore methylation biomarkers associated with the survival of various human cancers (25). MethSurv is freely available at https://biit.cs.ut.ee/methsurv. Through the MethSurv website, we will analyze the DNA methylation analysis of the selected AML-related genes in the TCGA database.
Results
Identification of DEGs and DMGs in AML
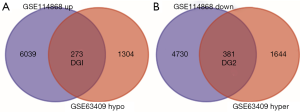
After obtaining DEGs and DMGs, 6,039 upregulated genes and 4,730 low-expression genes in GSE114868 were found from the gene expression microarray analysis. There were 1,644 hypermethylated genes and 1,304 hypomethylated genes in GSE63409 identified by the gene methylation microarray analysis. Further analysis of overlapping genes revealed 273 hypomethylated/upregulated genes and 381 hypermethylated/downregulated genes (Figure 2A,B).
KEGG pathway analysis
KEGG analysis was performed with hypomethylated/high-expression genes and hypermethylated/low-expression genes, and the first ten pathways were selected. Hypermethylated/low-expression genes were enriched in Th1 and Th2 cell differentiation, Epstein-Barr virus infection, Endocytosis, Th17 cell differentiation, Chemokine signaling pathway, Phagosome, Human immunodeficiency virus 1 infection, Toxoplasmosis, Human T-cell leukemia virus 1 infection, Human cytomegalovirus infection (Figure 3A). The hypomethylated/high-expression genes were significantly enriched in the thermogenesis, retrograde endocannabinoid signaling, oocyte meiosis, transcriptional misregulation in cancer, pathways in cancer, parathyroid hormone synthesis, secretion and action, glycine, serine and threonine metabolism, cholinergic synapse, Cushing syndrome, basal transcription factors (Figure 3B).

GO term analysis
GO analysis was performed with hypomethylated/high-expression genes and hypermethylated/low-expression genes, and the first ten pathways were selected. Regarding the biological processes (BP), the hypomethylated/upregulated genes were significantly enriched in regulation of methylation-dependent chromatin silencing, positive regulation of histone methylation, regulation of histone H3-K9 methylation, negative regulation of gene expression, epigenetic, L-serine metabolic process, regulation of intracellular steroid hormone receptor signaling pathway, negative regulation of small GTPase mediated signal transduction, regulation of Ras protein signal transduction, negative regulation of gene expression, glial cell development (Figure 4A). In the molecular function (MF), the hypomethylated/high-expression genes were enriched in helicase activity, proton-transporting ATP synthase activity, rotational mechanism, protein kinase binding, divalent inorganic cation transmembrane transporter activity, transcription coactivator activity, phosphatidylethanolamine binding, hyaluronoglucosaminidase activity, damaged DNA binding, S-adenosylmethionine-dependent methyltransferase activity, RNA binding (Figure 4B). In the cellular component (CC), the analysis revealed that enrichment mainly occurred at intrinsic component of the cytoplasmic side of the plasma membrane, nuclear speck, mitochondrial proton-transporting ATP synthase complex, mitochondrial matrix, mitochondrial proton-transporting ATP synthase complex, coupling factor F(o), cytoplasmic vesicle membrane, cytoplasmic ribonucleoprotein granule, cytoplasmic vesicle, nuclear body, Cul4A-RING E3 ubiquitin ligase complex (Figure 4C).
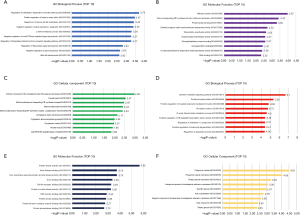
The BP enriched by the hypermethylated/low-expression genes included cytokine-mediated signaling pathway, protein phosphorylation, positive regulation of myeloid leukocyte mediated immunity, phosphorylation, N-acetylneuraminate metabolic process, positive regulation of NF-kappaB transcription factor activity, positive regulation of leukocyte degranulation, regulation of interleukin-12 production, regulation of apoptotic process, interferon-gamma-mediated signaling pathway (Figure 4D). In the MF, the hypomethylated/high-expression genes were enriched in protein kinase activity, actin filament binding, non-membrane spanning protein tyrosine kinase activity, actin-binding, Toll-like receptor binding, protein tyrosine kinase activity, PDZ domain binding, protein kinase binding, protein kinase A catalytic subunit binding, protein phosphatase binding (Figure 4E). In the CC, the hypermethylated/low-expression genes were enriched in the phagocytic vesicle, phagocytic vesicle membrane, tertiary granule membrane, is an integral component of endoplasmic reticulum membrane, specific granule, actin cytoskeleton, which is integral component of lumenal side of endoplasmic reticulum membrane, early endosome, tertiary granule, Golgi membrane (Figure 4F).
PPI network construction and module choice
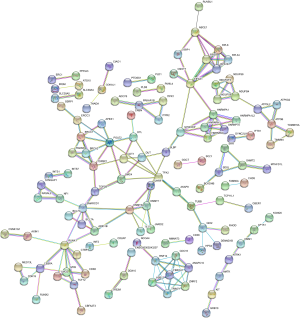
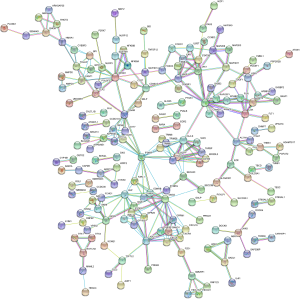
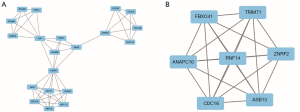
PPI network analysis was performed through the STRING database and Cytoscape software. We obtained a PPI network map of hypomethylation/upregulation genes and hypermethylation/downregulation genes. The PPI network map of the hypomethylated/upregulated gene has 256 nodes and 176 edges (Figure 5), and the PPI network map of the hypermethylated/downregulated gene has 373 nodes and 309 edges (Figure 6). The above two PPI network diagrams were respectively imported into the Cytoscape software to construct a module through the MCODE plug-in, in which the hypomethylation/upregulation gene was constructed into 9 modules, and the hypermethylation/downregulation gene was constructed into 12 modules. We selected the highest scoring modules (Figure 7A,B).
Survival analysis of genes
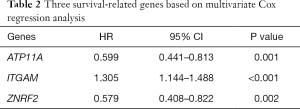
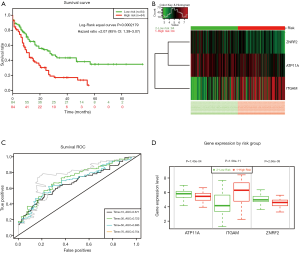
To assess whether the identified prognostic markers are valuable for predicting patient survival, we focused on the genes in the highest-scoring modules. We used SurvExpress software to analyze hypomethylated/upregulated, and hypermethylated/downregulated genes for univariate and multivariate Cox regression analysis. AML samples were divided into two groups: (I) high-risk group; (II) low-risk group. First, we performed the univariate Cox regression analysis of the highest-scoring module gene. The hypermethylation/downregulation gene module 1 has 20 genes, and the hypomethylation/upregulation gene module 1 contains 7 genes (Table 1). We screened for genes with P values <0.05 (ATP11A, ITGAM, ZNRF2). These three genes were then subjected to multivariate Cox regression analysis and found to have P values <0.05 (Table 2). The three genes were independent prognostic factors, and then we use three genes to establish a prognostic model (Figure 8A,B). The ROC curves assessed the accuracy of the prognostic model at 10, 30, 50, and 70 months (Figure 8C). These three genes were used to evaluate the prognosis of AML cytogenetic risk typing and cell morphological typing and found that these three genes differed in the intermediate/Normal and M2 prognostic analysis (Table 3). Finally, we show the expression levels and risk assessment of three key genes (Figure 8D).

Full table
Full table
Full table
DNA methylation data in MethSurv
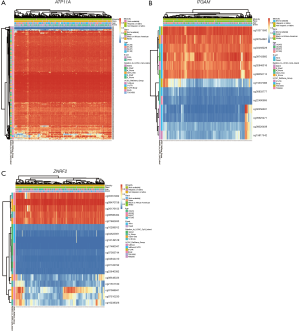
MethSurv is used to explore methylation biomarkers associated with the survival of various human cancers. MethSurv analyzed DNA methylation in TCGA. We performed methylation analysis of three genes (ATP11A, ITGAM, ZNRF2) that can be used as independent prognostic factors for AML in SurvExpress software. In the MethSurv software, we found that the P value of 73 CpG sites in the hypermethylated/down-regulated ATP11A in AML was <0.05, which we considered statistically significant. The top ten sites were shown in Table 4. Similarly, the hypermethylation/downregulation gene ITGAM has two CpG sites with a P value of <0.05, and the hypomethylation/upregulation gene ZNRF2 has three CpG sites with a P value of <0.05, which is statistically significant (Table 4). We found that the difference in DNA methylation between cg00990020 of ATP11A, cg05625471 of ITGAM, and cg07510230 of ZNRF2 was most pronounced (Figure 9A,B,C).
Full table
Discussion
AML is the most common acute leukemia in adults and is a highly heterogeneous and fatal (1). DNA methylation is the most studied epigenetic modification and is essential in the promotion of many important BP (7). Abnormal DNA methylation can lead to a variety of pathological conditions, including carcinogenesis. (8) Certain DNA methylation engages in the initial stages of carcinogenesis, such as RASSF1A in ovarian cancer (26). In addition, DNA methylation is stable over some time and can be detected non-invasively in the blood (27). Therefore, DNA methylation has the exciting potential to become an early diagnostic biomarker for cancer. Multiple studies have shown that gene expression abnormalities are closely related to AML (10). Identification of novel biomarkers will aid in early diagnosis and improved prognosis. In this study, we performed a comprehensive analysis of DNA methylation and gene expression to identify key epigenetic genes in AML.
This study is the first to report bioinformatics studies and its association between gene methylation of AML and the corresponding mRNA expression. Through our research, we found three genes that can be used as independent prognostic factors for AML. These three genes are the low expression/methylation genes ATP11A and ITGAM, and the high expression/low methylation gene ZNRF2. ATP11A is an adenosine triphosphate binding cassette (ABC) transporter homolog gene and belongs to an extended family of ABC transporters that confer multidrug resistance to cancer cells. For example, in lymphocytic leukemia, cancer cells are resistant by increasing ATP11A expression (28). In previous studies, it was found that the expression level of ATP11A gene in colorectal cancer tumor tissues was significantly higher than that in corresponding normal tissues, and it was important for the prognosis evaluation of colorectal cancer (29). Studies have shown that the ATP11A gene is a methylation biomarker for prostate cancer and is expressed in patients with metastatic and lethal PCA (30). ITGAM is a major non-human leukocyte antigen associated with the pathogenesis of autoimmune diseases such as systemic lupus erythematosus (SLE) and IgA nephropathy (31). Recent reports that SNP rs4597342 in ITGAM 3'UTR affect miR-21 binding may be considered a risk factor for psoriasis development (32). However, the above two genes have not been reported in AML. ZNRF2 is a ubiquitin ligase of the RING superfamily. It has been shown that membrane-associated E3 ubiquitin ligase ZNRF2 is involved in mTor activation and regulation through protein interactions, and ZNRF2 depletion reduces cell size and cell proliferation (33). ZNRF2 also plays a crucial role in tumorigenesis. For example, ZNRF2 enhances the mTor and its downstream targets CyclinD1 and CDK in NSCLC cells, and the negative correlation between ZNRF2 and miR-100 in osteosarcoma specimens, low miR-100 is associated with poor prognosis in OS patients (34,35). To date, the role of the ZNRF2 gene in AML and how it regulates AML through aberrant methylation is unclear. These three genes may be good indicators for assessing the prognosis of AML. We used these three genes to set up an independent prognostic model with high accuracy, which can be used to assess the prognosis of patients with AML and as a good target for AML treatment.
This study had several limitations: (I) the small number of cases evaluated; (II) the results of the study have not been validated on clinical samples.
In summary, our research identified many aberrantly expressed genes and pathways that can be regulated by aberrant methylation in AML through a comprehensive analysis of gene expression and methylation microarrays. We identified some new markers and pathways through multi-database analysis that could be an accurate diagnosis and treatment for AML; however, determining the role of these identified genes in the AML process requires more research.
Acknowledgments
Funding: This study was supported by grants from the National Natural Science Foundation of China (81874049, 81602179, 81570198); the National Science and Technology Major Project for New Drug (No. 2017ZX301033); the Co-construction of Provincial and Department Project (WKJ-ZJ-1919); the Zhejiang Provincial Natural Science Foundation of China (LY19H160036). This study was supported by the Key Laboratory of Tumor Molecular Diagnosis and Individualized Medicine of Zhejiang Province, Zhejiang Provincial People’s Hospital (People’s Hospital of Hangzhou Medical College). Hangzhou Health Science and Technology Project (No. Y201869156).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work, and the questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Nagler E, Xavier MF, Frey N. Updates in immunotherapy for acute myeloid leukemia. Transl Cancer Res 2017;6:86-92. [Crossref]
- Lai C, Doucette K, Norsworthy K. Recent drug approvals for acute myeloid leukemia. J Hematol Oncol 2019;12:100. [Crossref] [PubMed]
- Xu P, Wang M, Jiang Y, et al. The association between expression of hypoxia inducible factor-1α and multi-drug resistance of acute myeloid leukemia. Transl Cancer Res 2017;6:198-205. [Crossref]
- Vasu S, Kohlschmidt J, Mrózek K, et al. Ten-year outcome of patients with acute myeloid leukemia not treated with allogeneic transplantation in first complete remission. Blood Adv 2018;2:1645-50. [Crossref] [PubMed]
- Koutsi A, Vervesou EC. Diagnostic molecular techniques in haematology: recent advances. Ann Transl Med 2018;6:242. [Crossref] [PubMed]
- Kramer A, Challen GA. The epigenetic basis of hematopoietic stem cell aging. Semin Hematol 2017;54:19-24. [Crossref] [PubMed]
- Kulis M, Esteller M. DNA methylation and cancer. Adv Genet 2010;70:27-56. [Crossref] [PubMed]
- Yang X, Wong MPM, Ng RK. Aberrant DNA Methylation in Acute Myeloid Leukemia and Its Clinical Implications. Int J Mol Sci 2019. [Crossref] [PubMed]
- Heath EM, Chan SM, Minden MD, et al. Biological and clinical consequences of NPM1 mutations in AML. Leukemia 2017;31:798-807. [Crossref] [PubMed]
- Ferrara F, Palmieri S, Leoni F. Clinically useful prognostic factors in acute myeloid leukemia. Crit Rev Oncol Hematol 2008;66:181-93. [Crossref] [PubMed]
- Hillert LK, Bettermann-Bethge K, Nimmagadda SC, et al. Targeting RIPK1 in AML cells carrying FLT3-ITD. Int J Cancer 2019;145:1558-69. [Crossref] [PubMed]
- Chen PY, Chen YT, Gao WY, et al. Nobiletin Down-Regulates c-KIT Gene Expression and Exerts Antileukemic Effects on Human Acute Myeloid Leukemia Cells. J Agric Food Chem 2018;66:13423-34. [Crossref] [PubMed]
- Singh AA, Mandoli A, Prange KH, et al. AML associated oncofusion proteins PML-RARA, AML1-ETO and CBFB-MYH11 target RUNX/ETS-factor binding sites to modulate H3ac levels and drive leukemogenesis. Oncotarget 2017;8:12855-65. [Crossref] [PubMed]
- Mill CP, Fiskus W, DiNardo CD, et al. RUNX1-targeted therapy for AML expressing somatic or germline mutation in RUNX1. Blood 2019;134:59-73. [Crossref] [PubMed]
- Hunter AM, Sallman DA. Current status and new treatment approaches in TP53 mutated AML. Best Pract Res Clin Haematol 2019;32:134-44. [Crossref] [PubMed]
- Monma F, Nishii K, Shiga J, et al. Detection of the CBFB/MYH11 fusion gene in de novo acute myeloid leukemia (AML): a single-institution study of 224 Japanese AML patients. Leuk Res 2007;31:471-6. [Crossref] [PubMed]
- Mohamed AM, Balsat M, Koering C, et al. TET2 exon 2 skipping is an independent favorable prognostic factor for cytogenetically normal acute myelogenous leukemia (AML): TET2 exon 2 skipping in AML. Leuk Res 2017;56:21-8. [Crossref] [PubMed]
- Spencer DH, Russler-Germain DA, Ketkar S, et al. CpG Island Hypermethylation Mediated by DNMT3A Is a Consequence of AML Progression. Cell 2017;168:801-816.e13. [Crossref] [PubMed]
- Faderl S, Ferrajoli A, Harris D, et al. WP-1034, a novel JAK-STAT inhibitor, with proapoptotic and antileukemic activity in acute myeloid leukemia (AML). Anticancer Res 2005;25:1841-50. [PubMed]
- Du W, Lu C, Zhu X, et al. Prognostic significance of CXCR4 expression in acute myeloid leukemia. Cancer Med 2019;8:6595-603. [Crossref] [PubMed]
- Shah MV, Barochia A, Loughran TP. Impact of genetic targets on cancer therapy in acute myelogenous leukemia. Adv Exp Med Biol 2013;779:405-37. [Crossref] [PubMed]
- Chen SL, Qin ZY, Hu F, et al. The Role of the HOXA Gene Family in Acute Myeloid Leukemia. Genes 2019. [Crossref] [PubMed]
- Liang P, Hu R, Liu Z, et al. NAT10 upregulation indicates a poor prognosis in acute myeloid leukemia. Curr Probl Cancer 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One 2013;8:e74250. [Crossref] [PubMed]
- Modhukur V, Iljasenko T, Metsalu T, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018;10:277-88. [Crossref] [PubMed]
- Si JG, Su YY, Han YH, et al. Role of RASSF1A promoter methylation in the pathogenesis of ovarian cancer: a meta-analysis. Genet Test Mol Biomarkers 2014;18:394-402. [Crossref] [PubMed]
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer 2003;3:253-66. [Crossref] [PubMed]
- Zhang B, Groffen J, Heisterkamp N. Resistance to farnesyltransferase inhibitors in Bcr/Abl-positive lymphoblastic leukemia by increased expression of a novel ABC transporter homolog ATP11a. Blood 2005;106:1355-61. [Crossref] [PubMed]
- Miyoshi N, Ishii H, Mimori K, et al. ATP11A is a novel predictive marker for metachronous metastasis of colorectal cancer. Oncol Rep 2010;23:505-10. [PubMed]
- Zhao S, Geybels MS, Leonardson A, et al. Epigenome-Wide Tumor DNA Methylation Profiling Identifies Novel Prognostic Biomarkers of Metastatic-Lethal Progression in Men Diagnosed with Clinically Localized Prostate Cancer. Clin Cancer Res 2017;23:311-9. [Crossref] [PubMed]
- Fan Y, Li LH, Pan HF, et al. Association of ITGAM polymorphism with systemic lupus erythematosus: a meta-analysis. J Eur Acad Dermatol Venereol 2011;25:271-5. [Crossref] [PubMed]
- Hruska P, Kuruczova D, Vasku V, et al. MiR-21 binding site SNP within ITGAM associated with psoriasis susceptibility in women. Plos One 2019;14:e0218323. [Crossref] [PubMed]
- Hoxhaj G, Caddye E, Najafov A, et al. The E3 ubiquitin ligase ZNRF2 is a substrate of mTORC1 and regulates its activation by amino acids. Elife 2016. [Crossref] [PubMed]
- Xiao Q, Yang Y, An Q, et al. MicroRNA-100 suppresses human osteosarcoma cell proliferation and chemo-resistance via ZNRF2. Oncotarget 2017;8:34678-86. [PubMed]
- Zhang XF, Guo ZQ, Zhao CC, et al. The role of ZNRF2 in the growth of non-small cell lung cancer. Eur Rev Med Pharmacol Sci 2016;20:4011-7. [PubMed]