
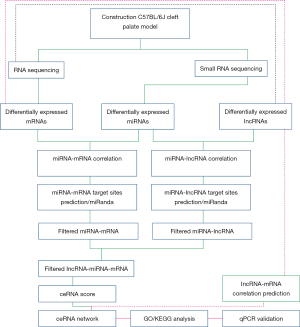
Integrated analysis identifying long non-coding RNAs (lncRNAs) for competing endogenous RNAs (ceRNAs) network-regulated palatal shelf fusion in the development of mouse cleft palate
Introduction
Cleft palate (CP) results from failure of embryonic palate shelf apposition and fusion at the midline during embryogenesis (1) and is considered to be caused by the individual and combined effects of genetic and environmental factors (2,3). Epithelial-mesenchymal transition (EMT) of the medial-edge epithelium/midline epithelial seam (MEE/MES) is a crucial process of palatal shelf fusion during palatogenesis (4,5). Thus, any imbalance in MEE/MES cell proliferation and apoptosis, or dysfunction of EMT, may lead to CP formation during palatal shelf fusion (6). In mice, the embryonic palatal shelf first grows vertically in the oral cavity from embryonic gestation day 12.5 (E12.5) to E14; then, at E14.5, the palatal shelves are elevated to the horizontal position and contact each other to complete initial palatal fusion (6). However, the detailed mechanisms regulating palatogenesis remain unclear. Although microRNAs (miRNAs) and/or long non-coding RNAs (lncRNAs) are increasingly being recognized as playing important roles in the development and pathogenesis of several diseases, the regulatory mechanisms by which miRNAs and/or lncRNAs directly or indirectly interact during palatal shelf fusion remain elusive.
Salmena’s research group recently proposed the competing endogenous RNA (ceRNA) hypothesis, which suggests that lncRNAs can “sponge” miRNAs to impair their activity by using shared miRNA response elements (MREs), resulting in the upregulated expression of target genes (7). Subsequent studies have also demonstrated that lncRNAs contain MREs and can act as ceRNAs to compete with mRNAs for the same binding sites on miRNA, consequently influencing multiple biological processes. For example, lncRNA H19 acts as a ceRNA for miR-138 and miR-200a to modulate the expression of multiple genes involved in the EMT (8). Liu et al. (9) further showed that the AEG-1 3'-untranslated region (UTR) functions as a ceRNA to induce EMT in human non-small cell lung cancer cells through the regulation of miR-30a activity. Nevertheless, how lncRNAs, when acting as ceRNAs or miRNA sponges, regulate miRNA in palatal fusion remains unknown.
To clarify this issue, the current study conducted high-throughput sequencing on RNAs and small RNAs obtained from CP embryos of pregnant C57BL/6J mice treated with all-trans retinoic acid (ATRA) (10), which were then compared to control embryos. We analyzed the aberrantly expressed RNA sequencing profiles for the ceRNA hypothesis to identify a new ceRNA network associated with the development of CP.
Methods
Animals and treatment
The flow chart for the experimental design is displayed in Figure 1. C57BL/6J mice (20–28 g body weight and 8–10 weeks of age; Beijing Vital River Laboratory Animal Technology, Beijing, China) were fed in the Translational Medicine Laboratory of the Second Affiliated Hospital of Shantou University Medical College (Shantou, China). Pregnant females at E10.5 were randomly divided into two groups: a control group and an ATRA-treated group. At E14.5, mice were euthanized, and palatal shelves were dissected using microbiological instruments. Palatal shelf tissues were stored as reported previously (10).
Histomorphology analysis
After palatal shelf tissues of the ATRA-untreated and ATRA-treated embryos (E14.5) were harvested, they were immediately photographed using a Leica M8 digital imaging system camera for morphological observation. The palatal shelf tissues were then embedded in paraffin and sectioned in either the coronal plane. After this, 4 µm paraffin sections were stained with hematoxylin and eosin (H&E) for morphological analysis.
Small RNA-seq and aberrantly expressed RNA profile analysis
Total RNA for miRNA sequencing was extracted from embryonic palatal shelf tissues (n=6, 3 ATRA-treated samples vs. 3 control samples) and a small RNA library was created with TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. The library quality was checked with an Agilent 2100 Bioanalyzer, and 50-bp single-end sequencing was performed according to the instructions of the Illumina HiSeq X platform (Illumina, San Diego, CA, USA). Reads without a 3' adapter and insert tag, and those shorter than 15 nt and longer than 41 nt were filtered from the raw data, and the clean reads were obtained. The miRNA expression level in both the ATRA-exposed group and the control group were assessed for the 3 biological and 3 technical replicates.
For the primary analysis, these RNAs were aligned and then subjected to a BLAST (11) search against Rfam v.10.1 (http://www.sanger.ac.uk/software/Rfam) (12) and GenBank databases (http://www.ncbi.nlm.nih.gov/genbank/). The known miRNAs were identified by aligning against the miRBase v.21 (http://www.mirbase.org/) (13). Based on the hairpin structure of a pre-miRNA and miRBase database, the corresponding miRNA star sequence was also identified. Aberrantly expressed miRNAs were identified (P<0.05) and predicted using miRanda software (14), under the following parameters: S ≥150 ΔG ≤−30 kcal/mol and strict demand strict 5' seed pairing.
RNA-seq and differential expression profile analysis
Total RNA in the embryonic palatal shelf tissue (n=6, 3 ATRA-treated samples vs. 3 control samples) was isolated with TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Libraries were constructed using TruSeq Stranded Total RNA with Ribo-Zero Gold (Illumina Inc., San Diego, CA, USA) according to the manufacturer’s protocol. Raw sequencing read data was stored in FastQC (v0.11.4). After removing the reads that contained the adapter contamination (i.e., low-quality bases and undetermined bases), the high-quality paired-end reads for each sample were mapped to the mouse reference genome (
Full table
Full table
Correlation analysis
To compare the expression level of a gene across different samples (n=6, 3 ATRA-treated samples vs. 3 control samples), read counts obtained from the RNA‐seq data were normalized as FPKM. For each RNA, we calculated the Pearson correlation coefficient of its expression level for that of each differentiated miRNA/lncRNA/mRNA. A correlation coefficient between miRNA and lncRNA/miRNA and mRNA/lncRNA and mRNA smaller than 0.05 with an absolute value of correlation greater than 0.7 was considered to have potential relevance.
miRNA target prediction
The miRNA target prediction process was as follows. First, the miRNA-target relationship pair prediction was performed with the base sequence using miRanda (14) to identify the miRNA-binding sites, including the position on the miRNA and DNA sequence, and the free energy of MREs (14,16). In addition, the related lncRNA-mRNA pairs were identified to predict the miRNA-lncRNA/miRNA-mRNA pairs, and pairs that shared the same miRNA were identified as candidate lncRNA-miRNA-mRNA competing for interactions. Second, related miRNA-target pairs were predicted using co-expression values. The miRNA-lncRNA/miRNA-mRNA pairs were calculated, and the negative correlations of miRNA-target expression were screened out. Third, the overlapping miRNA-target pairs based on the results of intersection that were obtained from the first step (the miRNA-target relationship pair prediction using miRanda) and the second step (the related miRNA-target pairs using co-expression values) were filtered to improve the reliability of the miRNA-target pairs. The regulatory miRNA-target pairs were used for the subsequent prediction of ceRNAs.
Prediction of ceRNAs
If lncRNA can regulate miRNA and the miRNA can regulate an mRNA, the lncRNA may act as a ceRNA. According to the miRNA-target pair prediction, the prediction of the ceRNAs process was as follows. First, ceRNAs were predicted using ceRNA scores, which were calculated as reported previously (17,18). Second, ceRNAs were predicted using expression levels. We selected the ceRNA-related pairs according to the expression levels of the lncRNA-mRNA and miRNA-target pairs (19). Third, the results of the ceRNA score and expression levels were integrated to predict the ceRNA.
Functional prediction
In order to predict the ceRNA candidate genes involved in biological processes and signaling pathways, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses to assess its functions. For the coding genes with a ceRNA relationship, the hypergeometric distribution test was used to calculate the significance of differential gene enrichment in each GO or KEGG term. The calculated result returned a P value of enrichment significance, and a P value <0.05 indicated that differential gene enrichment occurred in the GO or KEGG term.
Construction of the ceRNA network
According to the prediction of ceRNAs, shared miRNA numbers were sorted from high to low, and pairs with fewer than 3 shared miRNAs were filtered out (20). Then, ceRNA networks were constructed for embryo palatal shelves from both control and ATRA-treated mice.
Quantitative polymerase chain reaction (qPCR) validation
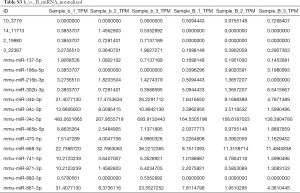
To validate the RNA-seq data, qPCR was conducted using 6 individual samples, 3 control, and 3 ATRA-treated. Each sample was subjected to 3 technical replications for qPCR. The qPCR primers used in this study are shown in Table 1. The miRNA, lncRNA, and mRNA expression levels were analyzed as described previously (21).

Full table
Statistical analysis
All statistical analyses were performed using SPSS 16.0 statistical software (SPSS, Chicago, IL, USA). The differential gene expression levels were assessed using the R package edgeR, as described previously (22). qPCR data were analyzed using Student’s t-test. Pearson correlation analysis was used in miRNA-target co-expression analyses.
Results
Histomorphology observations
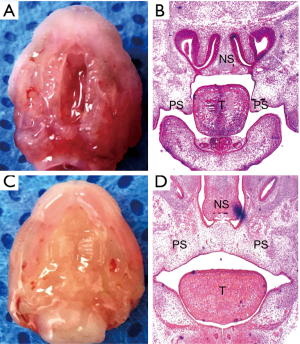
In this study, 20 pregnant C57BL/6J mice were examined, and 142 embryos were collected, including 86 embryos from the ATRA-treated group, and 56 embryos from the control group. Each pregnant mouse had multiple embryos. There was at least one CP embryo in each pregnant mouse in the ATRA-treated group, while there were no CP embryos in the control group. The incidence of CP in the ATRA-treated group was 100%. At E14.5, the ATRA-treated palatal shelves remained separated without fusion (Figure S1A,B). At the same time, the palatal control shelves had contacted each other to complete initial palatal fusion (Figure S1C,D).
Data analysis
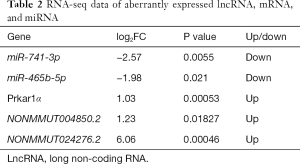
A total of 18 aberrantly expressed miRNAs (Table S3), 861 mRNAs (table online: http://fp.amegroups.cn/cms/7c8698af7959c572ba3f4bce0b9c1a65/atm.2019.11.93-1.pdf), and 583 lncRNAs (table online: http://fp.amegroups.cn/cms/aa6681b0fd4cd9036c06135d37d28842/atm.2019.11.93-2.pdf) were identified from the sequencing data. Among the aberrantly expressed lncRNAs, mRNAs, and miRNAs, NONMMUT004850.2 (P=0.01827, log2FC: 1.23), NONMMUT024276.2 (P=0.00046, log2FC: 6.06), and Prkar1α (P=0.00053, log2FC: 1.03) were up-regulated, respectively, whereas miR-741-3p (P=0.0055, log2FC: −2.57) and miR-465b-5p (P=0.021, log2FC: −1.98) were down-regulated (Table 2).
Full table
Full table
miRNA-target relationships predicted by correlation analysis
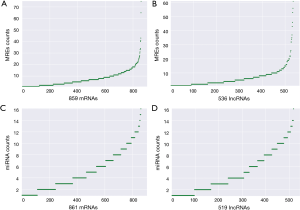
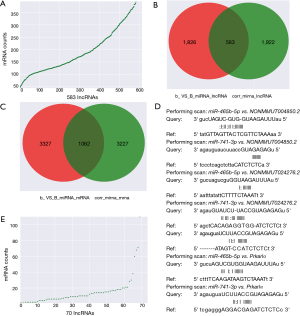
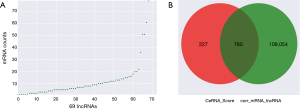
According to the obtained sequences, we identified a total of 6,251 MREs for 18 miRNAs that target 859 mRNAs (Figure 2A, table online: http://fp.amegroups.cn/cms/aa657847352490aa1cebb75a80959d00/atm.2019.11.93-3.pdf), and 3,312 MREs for 18 miRNAs that target 536 lncRNAs (Figure 2B, table online: http://fp.amegroups.cn/cms/84c11d0428a3f0234943a75cf2c5402c/atm.2019.11.93-4.pdf). Moreover, we identified a total of 4,839 interactions of 18 miRNAs with 861 mRNAs according to the co-expression values (Figure 2C). There were 2,505 interactions of 18 miRNAs with 519 lncRNAs (Figure 2D) and 109,834 interactions of 583 lncRNAs, with 395 of the identified mRNAs (Figure 3A).
The correlation analysis of NONMMUT004850.2/NONMMUT024276.2-miR-741-3p/miR-465b-5p-Prkar1α Pearson’s correlation coefficient showed that Prkar1α vs. NONMMUT004850.2/NONMMUT024276.2 is a positive correlation, and miR-741-3p/miR-465b-5p vs. NONMMUT004850.2/NONMMUT024276.2/Prkar1α is a negative correlation (Table 3). Pearson’s correlation coefficient’s absolute value >0.8 with a P value <0.05 was considered statistically significant. Furthermore, using miRanda, we obtained 583 interactions of 2,409 miRNA-lncRNA target pairs with 2,505 miRNA-lncRNA co-expression profiles (Figure 3B, table online: http://fp.amegroups.cn/cms/514ebd18cdb42cb898b8b78417f8e5bb/atm.2019.11.93-5.pdf), and 1,062 interactions of 4,389 miRNA-mRNA target pairs with 4,289 miRNA-mRNA co-expression profiles (Figure 3D, table online: http://fp.amegroups.cn/cms/152d8707844d0b419baf806cf32aa70c/atm.2019.11.93-6.pdf). Among the miRNA-target-binding sites, miR-741-3p/miR-465b-5p-target binding NONMMUT024276.2/NONMMUT004850.2 and the Prkar1α 3'-UTR are shown in Figure 3D.

Full table
Constructing the ceRNA network
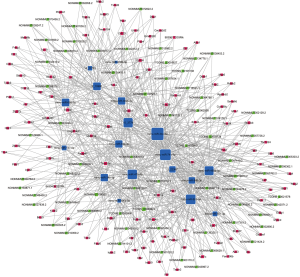
According to the ceRNA scores, we obtained 1,007 ceRNAs (lncRNA-miRNA-mRNA) for 70 lncRNAs with 112 mRNAs (Figure 3E, table online: http://cdn.amegroups.cn/static/application/7a591ed630a25eef64f4ba47aa81d058/atm.2019.11.93-7.pdf). Based on the co-expression values, 799 ceRNAs (lncRNA-miRNA-mRNA) of 69 lncRNAs with 78 mRNAs (Figure 4A, table online: http://cdn.amegroups.cn/static/application/eb89017efa20b79eb35fa935024c91a2/atm.2019.11.93-8.pdf) were obtained. We then computed the results of the ceRNA scores and further percolated and excluded the overlapped ceRNA. In total, 780 ceRNAs between 1,007 ceRNA scores and 109,834 lncRNA-mRNA pairs were identified (Figure 4B). The ceRNA network was then constructed based on 69 lncRNAs, 18 miRNAs, and 78 mRNAs using Cytoscape 3.5.1 (23) (Figure 5).
GO and KEGG analysis
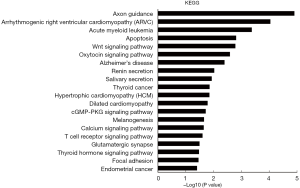
GO analyses for biological process showed that the ceRNA-related genes were involved in “transcription from RNA polymerase II promoter”, “positive regulation of transcription, DNA-templated”, “gene silencing”, “face morphogenesis”, and “neural tube closure” (Figure 6). Prkar1α was found to be involved in “regulation of protein kinase activity” and “negative regulation of cyclic AMP (cAMP)-dependent protein kinase activity” (table online: http://fp.amegroups.cn/cms/0a8ef22d1452ed151a738bcb6faf482c/atm.2019.11.93-9.pdf). Moreover, KEGG analyses showed that the ceRNA-associated genes were involved in “apoptosis”, “Wnt signaling pathway”, and “Focal adhesion” (Figure 7). The KEGG database showed an association of Prkar1α with “apoptosis” (table online: http://fp.amegroups.cn/cms/b4e6f7b0a030c020405ad82e0a9527d2/atm.2019.11.93-10.pdf) (P<0.05, fold enrichment >2).

qPCR validation
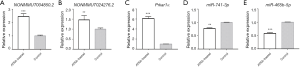
qPCR was performed to further validate NONMMUT004850.2, NONMMUT024276.2, Prkar1α, and miR-741-3p/miR-465b-5p expression levels between the groups. The results showed that NONMMUT004850.2 (P=5E−05), NONMMUT024276.2 (P=0.0012), and Prkar1α (P=3E−05) were up-regulated, whereas miR-741-3p (P=0.006)/miR-465b-5p (P=1E−04) was down-regulated (Figure 8). Therefore, the qPCR change trend was in concordance with the sequencing profiling.
Discussion
The ceRNA hypothesis proposes a novel molecular, multiple-level gene regulation mechanism that figures significantly in pathological processes (7). However, our understanding of the molecular mechanisms that allow lncRNAs to act as ceRNAs in the palatogenesis of CP is still limited. In our study, we systematically analyzed sequencing data between E14.5 embryos from control and ATRA-treated mice as a CP model. Bioinformatics analyses of the aberrantly expressed RNAs between the groups led us to speculate that the NONMMUT004850.2/NONMMUT024276.2-miR-741-3p/miR-465b-5p-Prkar1α ceRNA network may be associated with the development of CP.
Prkar1α is located at chromosome 17q24 and encodes a regulatory subunit of protein kinase A (PKA) that regulates neural crest cell formation, migration, and differentiation during embryonic development (24). Embryonic cranial neural crest (CNC) cells undergo complex molecular and morphological changes in order to form the vertebrate skull, and nearly three quarters of all birth defects, including CP, result from defects in craniofacial development (15,25). cAMP-mediated growth control in embryonic palatal development may be further modulated by endogenous prostaglandin and/or catecholamine, which depresses palatal cAMP levels (26,27). Previous studies show that cAMP levels increase before palatal fusion, which is coupled with increased PKA activation. In addition, PKA activity decreases to baseline after palatal fusion (26,27). In the present study, we discovered that NONMMUT004850.2/NONMMUT024276.2 acts as a ceRNA for miR-741-3p/miR-465b-5p to modulate the expression of Prkar1α, which results in the negative regulation of PKA activity, thereby blocking palatal fusion.
miRNA-induced target gene silencing requires target gene recognition by the miRNA-loaded RNA-induced silencing complex (RISC); however, sequence complementarity between miRNAs and their targets is rare in animals (28). Instead, complementarity between the miRNA seed sequence and the target gene is sufficient for target recognition (29). Based on the ceRNA hypothesis, the NONMMUT004850.2/NONMMUT024276.2-miR-741-3p/miR-465b-5p-Prkar1α ceRNA networks are supported by the following evidence. First, the prediction of the miR-741-3p/miR-465b-5p-MREs showed that miR-741-3p/miR-465b-5p vs. NONMMUT024276.2/NONMMUT004850.2, and miR-741-3p/miR-465b-5p vs. Prkar1α have sufficient sequence complementarity (Figure 3D. Second, the ceRNA network was constructed by multiple filtration methods involving ceRNA-related lncRNAs, miRNAs, and mRNAs. Third, functional annotation indicated that Prkar1α is involved in “regulation of protein kinase activity”, “negative regulation of cAMP-dependent protein kinase activity”, and “apoptosis” pathways. Fourth, the NONMMUT004850.2/NONMMUT024276.2-miR-741-3p/miR-465b-5p-Prkar1α network is located at the ceRNA network hub (Figure 5). Finally, the qPCR results showed that NONMMUT004850.2, NONMMUT024276.2, miR-741-3p/miR-465b-5p, and Prkar1α change in concordance with the sequencing profiling.
Despite our findings, it should be noted that our study is still preliminary in nature, and thus further research is needed to completely elucidate the ceRNA-regulation relationship of gene alterations occurring during CP formation. In addition, our sample size is relatively small, and the palatal shelves have been obtained directly from embryonic tissues. Therefore, it is possible that the target tissue could have been contaminated with samples of surrounding tissue. Another limitation is that we cannot exclude the effect of ATRA and its endogenous small molecule metabolites on the experimental results. We expect to integrate an analysis that identifies lncRNAs for a ceRNA network, which can help to further deepen our understanding of this level of lncRNA-dependent post-transcriptional regulation.
Conclusions
In summary, by constructing and functionally analyzing lncRNA-related networks based on the ceRNA hypothesis, we identified a NONMMUT004850.2/NONMMUT024276.2-miR-741-3p/miR-465b-5p-Prkar1α ceRNA network that is potentially associated with palatogenesis and the formation of CP, and thus merits further study.
Acknowledgments
Funding: This research was funded by the National Natural Science Foundation of China (grant No. 81571920).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The guidelines and protocols for these experiments were approved by the Ethical Committee of the Medical College of Shantou University, Shantou, Guangdong, China (SUMC2015-106).
References
- Rahimov F, Jugessur A, Murray J. Genetics of nonsyndromic orofacial clefts. Cleft Palate Craniofac J 2012;49:73-91. [Crossref] [PubMed]
- Dixon MJ, Marazita ML, Beaty TH, et al. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet 2011;12:167-78. [Crossref] [PubMed]
- Watkins SE, Meyer RE, Strauss RP, et al. Classification, epidemiology, and genetics of orofacial clefts. Clin Plast Surg 2014;41:149-63. [Crossref] [PubMed]
- Dudas M, Li W, Kim J, et al. Palatal fusion - where do the midline cells go? A review on cleft palate, a major human birth defect. Acta Histochem 2007;109:1-14. [Crossref] [PubMed]
- Iordanskaia T, Nawshad A. Mechanisms of transforming growth factor β induced cell cycle arrest in palate development. J Cell Physiol 2011;226:1415-24. [Crossref] [PubMed]
- Nawshad A. Palatal seam disintegration: to die or not to die? that is no longer the question. Dev Dyn 2008;237:2643-56. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 2015;6:22513-25. [Crossref] [PubMed]
- Liu K, Guo L, Guo Y, et al. AEG-1 3'-untranslated region functions as a ceRNA in inducing epithelial-mesenchymal transition of human non-small cell lung cancer by regulating miR-30a activity. Eur J Cell Biol 2015;94:22-31. [Crossref] [PubMed]
- Shu X, Shu S, Cheng H, et al. Genome-Wide DNA Methylation Analysis During Palatal Fusion Reveals the Potential Mechanism of Enhancer Methylation Regulating Epithelial Mesenchyme Transformation. DNA Cell Biol 2018;37:560-73. [Crossref] [PubMed]
- Altschul SF, Gish W, Miller W, et al. A basic local alignment search tool. 1990.
- Sam GJ, Alex B, Mhairi M, et al. Rfam: an RNA family database. Nucleic Acids Research 2003;31:439-41. [Crossref] [PubMed]
- Griffiths-Jones S, Saini HK, van Dongen S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res 2008;36:D154-8. [Crossref] [PubMed]
- Enright AJ, John B, Gaul U, et al. MicroRNA targets in Drosophila. Genome Biology 2003;5:R1. [Crossref] [PubMed]
- Jones GN, Pringle DR, Yin Z, et al. Neural crest-specific loss of Prkar1a causes perinatal lethality resulting from defects in intramembranous ossification. Mol Endocrinol 2010;24:1559-68. [Crossref] [PubMed]
- Liu K, Yan Z, Li Y, et al. Linc2GO: a human LincRNA function annotation resource based on ceRNA hypothesis. Bioinformatics 2013;29:2221-2. [Crossref] [PubMed]
- Ala U, Karreth F, Bosia C, et al. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc Natl Acad Sci USA 2013;110:7154-9. [Crossref] [PubMed]
- Das S, Ghosal S, Sen R, et al. lnCeDB: database of human long noncoding RNA acting as competing endogenous RNA. PLoS One 2014;9:e98965. [Crossref] [PubMed]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 2005;4:Article17.
- Tay Y, Kats L, Salmena L, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011;147:344-57. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 2011;696:291-303. [Crossref] [PubMed]
- Marazita ML, Mooney MP. Current concepts in the embryology and genetics of cleft lip and cleft palate. Clin Plast Surg 2004;31:125-40. [Crossref] [PubMed]
- Selleck MA, Bronner-Fraser M. Origins of the avian neural crest: the role of neural plate-epidermal interactions. Development 1995;121:525-38. [PubMed]
- Greene RM, Pratt RM. Correlation between cyclic-AMP levels and cytochemical localization of adenylate cyclase during development of the secondary palate. J Histochem Cytochem 1979;27:924-31. [Crossref] [PubMed]
- Greene RM. Signal transduction during craniofacial development. Crit Rev Toxicol 1989;20:137-52. [Crossref] [PubMed]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010;11:597-610. [Crossref] [PubMed]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20. [Crossref] [PubMed]


