
Integrated analysis and knockdown of RAB23 indicate the role of RAB23 in gastric adenocarcinoma
Introduction
Gastric cancer, a common and pernicious disease, is the third most common cause of cancer death worldwide. Morbidity and mortality differ geographically, with East Asia having the highest mortality and morbidity (1,2). The 5-year survival rate is more than 90% if the tumor is diagnosed at a very early stage and resected. Nevertheless, early stages of gas chromatography (GC) are usually asymptomatic, and if patients are diagnosed at advanced stage, the 5-year survival rate is reduced to 20% (3). Therefore, the development of early noninvasive biomarkers is essential for the early diagnosis of GC, which may improve the prognosis of GC.
Gastroscopy is widely used for early screening, but it is invasive, uncomfortable and expensive. Some valid ways to enhance prognosis are to identify early cancerous changes and metastases, to study the molecular mechanism of cancer occurrence, and to find biomarkers for the early diagnosis of GC. RAB23 is associated with the occurrence and metastasis of tumors (4). Recently, numerous studies have indicated that RAB23 is upregulated in renal cell carcinoma (5), hepatocellular carcinoma (6), osteosarcoma (7), esophageal squamous cell carcinoma (8) and bladder cancer (9), which are regulated by miR-384, miR-16, miR-802, miR-665 and miR-92b. According to our integrated microarray analysis, RAB23 is upregulated in the tissues of patients with gastric adenocarcinoma and involved in the PI3K-Akt signaling pathway and intracellular vesicle processes. Furthermore, RAB23 was a predicted target of hsa-miR-130a-3p and hsa-miR-363-3p and was dysregulated in the tissues of patients with gastric adenocarcinoma. To further research the roles of RAB23 in gastric adenocarcinoma, MKN45 cells were transfected with shRNA-RAB23 to inhibit the expression of RAB23.
Thus, we aimed to determine the expression of RAB23 in GC, analyze the association between RAB23 expression and tumor progression, and further explore the possible underlying mechanism. We found that RAB23 was elevated in GC, was related to the diagnosis of GC, and was regulated by hsa-miR-130a-3p and hsa-miR-363-3p and that RAB23 knockdown by shRNA interference reduced proliferation and migration and enhanced apoptosis by suppressing the activation of PI3K/Akt signaling in gastric cancer.
Methods
Identification of differentially expressed mRNAs (DEmRNAs) and miRNAs (DEmiRNAs)
Based on the GEO database, we downloaded mRNA data from 104 samples (75 tumor tissues and 29 normal tissues) and miRNA data from 79 samples, including 66 tumor tissues and 13 normal tissues. MetaMA is an R package that performs meta-analysis methods for microarrays. The method used to merge p values was the inverse normal method. Multiple comparisons correction was performed to obtain the values of the false discovery rate (FDR) with the Benjamini & Hochberg method (10).
Protein-protein interaction (PPI) network
Next, we established the PPI network by using Biological General Repository for Interaction Datasets (BioGRID) (11) and Cytoscape (12) on the top 50 DEmRNAs between tumor tissues and normal tissues. Proteins with a degree of ≥30 were defined as hub proteins of the PPI network.
Detection of DEmiRNAs
To further investigate the correlation between DEmiRNAs and DEmRNAs, we obtained DEmiRNA-DEmRNA interaction pairs (including downregulated DEmiRNA-upregulated DEmRNA pairs and upregulated DEmiRNA-downregulated DEmRNA pairs) in tumor tissues vs. normal tissues. Detection of the target gene was performed as previously described (13).
Functional annotation of target genes
To explore the biological function of the target genes, we used WebGestalt to analyze all the DEmRNAs and target genes identified via the GO functional classification and the KEGG biochemical pathway analyses (http://www.webgestalt.org/option.php) (14).
Survival analysis for RAB23
The prognostic value of RAB23 was analyzed using OncoLnc (http://www.oncolnc.org/). Patients were divided into high-level and low-level groups based on the median value of RAB23. The overall survival (OS) rates of the patients in the high-level and low-level RAB23 groups were evaluated using Kaplan-Meier analysis (gepia.cancer-pku.cn/detail.php?gene=&clicktag=survival).
Cell culture
To validate the effect of selected DEmiRNAs and DEmRNAs on gastric cancer cells in vitro, we obtained the human gastric cancer cell line MKN45 from the Key Laboratory of Digestive Tract Malignancy, Ministry of Education of Fujian Medical University (Shanghai, China). Cells were transfected with shRNA-RAB23 (RAB23 knockdown, RAB23-KD) or shRNA negative control (NC). Cells with no treatment were used to determine background effects (the blank group).
Plasmid construction and transfection
Total RNA was isolated from MKN45 cells and reverse transcribed into complementary DNA (cDNA). The primer sequences of RAB23 were 5′‐TGACTGCGCACTTGGAATAGT‐3′ (forward) and 5′‐CATAACCACGGGGCCCTAAG‐3′ (reverse). The sequences of shRNA-7350, shRNA-7351, shRNA-7352 and shRNA-7353 were obtained from Oligobio. Cells were transfected with shRNA-7350, shRNA-7351, shRNA-7352 and shRNA-7353 or shRNA NC.
Cell proliferation, migration and apoptosis assays
Cell viability was detected using the Cell Counting Kit-8 (CCK-8) (Sigma-Aldrich, USA). We added 10 µL of the CCK8 reagent (Solarbio Science & Technology, Beijing, China) to every well, and then the cells were incubated for 90 min at 37 °C. We measured the OD value at 450 nm every 24 h (15).
We used transwell chambers to evaluate cell migration and invasion. Cells were digested with trypsin and resuspended in culture medium without serum, and approximately 1×104 cells were added to the upper chamber. Subsequently, the cells were incubated for 24 h, and the invasive cells were fixed using 4% paraformaldehyde and then stained with 0.1% crystal violet for 5 min. Finally, we imaged and counted the invasive cells.
Flow cytometry was utilized to evaluate cell apoptosis via the Annexin V-FITC-PI Apoptosis Detection Kit. Cells were analyzed via a FACSCalibur and a BD FACSDiva.
Quantitative reverse transcription PCR (RT-PCR) and Western blot
Using the SYBR Master Mixture, we detected the expression of mRNAs. Western blot assays were performed to detect the relative expression of RAB23, Bcl-2, Rac1 and PI3K. RIPA Lysis Buffer was utilized to lyse cells to extract proteins. This assay was performed as previously described (16).
Metabolomic analysis of gastric cancer cells using gas chromatography mass spectrometry (GC-MS)
To identify the influence of RAB23 on the metabolism of gastric cancer cells, we performed GC-MS analysis by using a GCMS-QP2010 Ultra with a DB-5MS capillary column. The temperature programming was as follows: the initial temperature of the column oven was 90 °C; the temperature was kept here for 0.2 min and then was increased by 10 °C/min to 180 °C, 5 °C/min to 240 °C, and 20 °C/min to 280 °C; the temperature was then held at 280 °C for 11 min. The raw data from the GC/MS experiments were imported into the Shimadzu GC solution software for analysis, including peak extraction, denoising, and deconvolution, and metabolites were characterized using the Fiehn Lab database.
Immunofluorescence
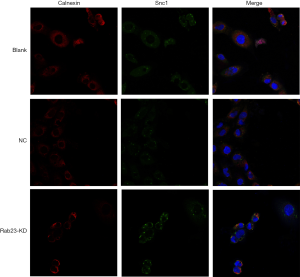
To assess the influence of RAB23 on vesicle transport, we performed colocalization analysis of Snc1, the vesicular protein VAMP3 and the endoplasmic reticulum protein Calnexin. Cells were fixed in 4% paraformaldehyde for 15 min and then incubated at 4 °C in PBS with 0.5% Triton X-100 for 3 min. Subsequently, 3% bovine serum albumin was used to block cells three times (5 min per time) at room temperature in PBS. We used immunofluorescence to detect protein expression at 4 °C overnight. Next, cells were incubated with secondary antibodies for 30 min at room temperature. Nuclei were counterstained with DAPI (Beyotime; Shanghai, China), and images were captured using an upright fluorescence microscope (DM3000; Leica, Solms, Germany) at room temperature (17).
Results
Identification of DEmRNAs and DEmiRNAs between gastric carcinoma tissues and adjacent normal tissues
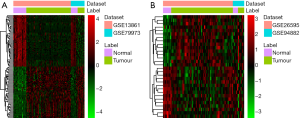
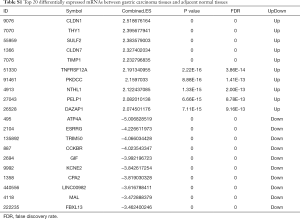
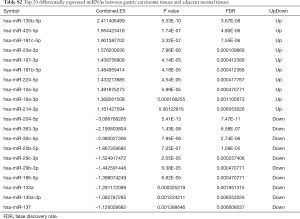
Two datasets (GSE79973 and GSE13861) of mRNAs and two datasets (GSE94882 and GSE26595) of miRNAs were used in our study. Compared with adjacent normal tissues, 4,586 DEmRNAs were identified in tumor tissues, including 2,606 upregulated DEmRNAs and 1,980 downregulated DEmRNAs. The top 100 upregulated and downregulated DEmRNAs are displayed in Figure S1A. Furthermore, a total of 30 DEmiRNAs (15 upregulated and 15 downregulated) were obtained. A hierarchical clustering map of DEmiRNAs is shown in Figure S1B. The details of the top 20 DEmRNAs and DEmiRNAs between gastric carcinoma tissues and adjacent normal tissues are shown in Tables S1,S2.
Full table
Full table
PPI network
The PPI network consisted of 586 nodes and 650 edges (Figure S2). Five hub proteins, including ENO1 (degree =69), HDGF (degree =66), CDK4 (degree =50), KPNA2 (degree =45) and ATP4A (degree =32), were identified according to the PPI network.
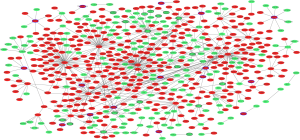
DEmiRNA-DEmRNA interaction network
A total of 5,091 DEmiRNA-DEmRNA pairs were obtained (Figure 1). Among these, 705 downregulated DEmiRNA-upregulated DEmRNA pairs and 592 upregulated DEmiRNA-downregulated DEmRNA pairs were predicted by ≥4 algorithms, and 667 validated downregulated DEmiRNA-upregulated DEmRNA pairs (Figure 1A) and 302 upregulated DEmiRNA-downregulated DEmRNA pairs (Figure 1B) were derived from miRWalk. Among these, hsa-miR-130a-3p (18) and hsa-miR-363-3p (19) were predicted to regulate the expression of RAB23 and were found to be related to gastric cancer after searching the literature. Therefore, RAB23 was selected as the focus of subsequent experiments.
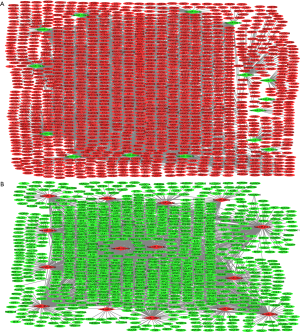
Functional annotation of DEmiRNA targets
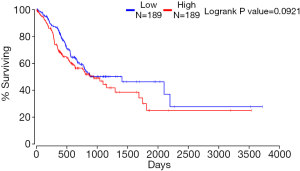
In Figure 2A,B,C, based on the GO enrichment analysis, cell proliferation (FDR =0), regulation of cell proliferation (FDR =2.85E-12), vesicle-mediated transport (FDR =0.003872877), endoplasmic reticulum (FDR =0), catalytic complex (FDR =5.72E-14), cytoplasmic vesicle (FDR =6.95E-12) and intracellular vesicle (FDR =6.95E-12) were the most significantly enriched GO terms in gastric cancer. The PI3K-Akt signaling pathway (FDR =0.002893618) and ECM-receptor interaction (FDR =0.002893618) were two significantly enriched pathways in gastric cancer (Figure 2D). TNFRSF10B (20) and HIPK2 (20,21) were enriched in the cell proliferation and regulation of cell proliferation terms. RAB23 (22) was enriched in the cytoplasmic vesicle and intracellular vesicle terms. The Kaplan-Meier survival curves of 378 GC cases showed that RAB23 was not significantly (P=0.0921) related to the prognosis of GC (Figure S3).

RAB23 knockdown inhibits the growth capacity and cell migration of gastric cancer in vitro
We transfected the gastric cancer cell line MKN45 with shRNA-7350, shRNA-7351, shRNA-7352 and shRNA-7353 or shRNA NC to inhibit the expression of RAB23. The expression level of RAB23 was significantly blocked by shRNA-7351 and shRNA-7353 in MKN45 cells (P<0.01, Figure 3A), and shRNA-7353 (RAB23 knockdown, RAB23-KD) was used for subsequent experiments.
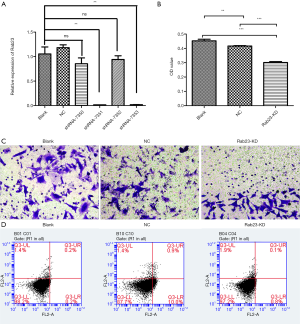
To examine the proliferative effect of RAB23 on gastric cancer cells, we utilized the gastric cancer cell line MKN45 for transfection with RAB23 shRNA-7353 to inhibit the expression of RAB23. Cell proliferation was determined by CCK8 assay. As presented in Figure 3B, the cell viability was decreased in RAB23-deficient MKN45 cells.
A transwell assay was performed to determine whether RAB23 affected tumor metastasis. As shown in Figure 3C, cell migration was slightly decreased in MKN45 cells. We observed that the RAB23-knockdown group did not have a changed apoptosis rate compared with the NC group or blank group (P>0.05, Figure 3D).
RAB23 knockdown inhibits the activation of the PI3K/Akt pathway in gastric cancer
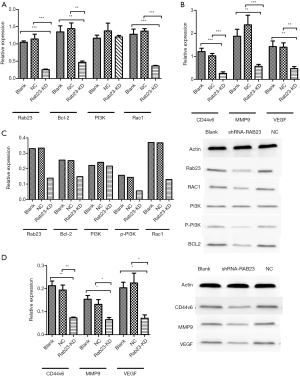
To investigate the mechanism by which RAB23 affects gastric cancer progression, we investigated the changes in the PI3K/Akt pathway after RAB23 knockdown in MKN45 cells. As the qPCR and Western blot results suggest, RAB23 knockdown significantly reduced the expression level of Bcl-2 and the tumor angiogenesis-promoting protein Rac1. There was no significant change in the level of PI3K protein or PI3K phosphorylation, RAB23 knockdown also significantly reduced the expression of CD44v6, MMP9 and VEGF (Figure 4). Additionally, immunofluorescence staining revealed that RAB23 influenced vesicle transport (Figure 5).
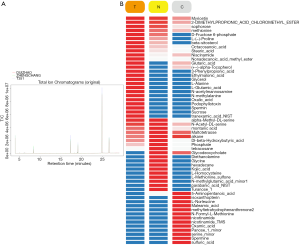
GC/MS metabolic profiling
Typical GC/MS TIC chromatograms of MKN45 cells with three different treatments and the matched normal control are shown in Figure 6A. As shown in Figure 6B, a total of 56 endogenous metabolites, including glycerol, niacinamide, and nonadecanoic acid methylester, were detected sequentially.
Discussion
Despite the development of diagnostic techniques and treatment methods for gastric adenocarcinoma, most patients are diagnosed with disease at an advanced stage (23). As a consequence, the 5-year survival rate is extremely poor, at <10%, and this diagnosis at an advanced stage is partially due to a lack of identified early-stage diagnostic markers and efficient monitoring methods for cancer progression. In the present study, after integrated analysis, RAB23 was indicated to be upregulated in gastric adenocarcinoma tissues compared to normal tissues, and this upregulation was affected by hsa-miR-130a-3p and hsa-miR-363-3p. We speculated that RAB23 may be a potential noninvasive biomarker for the early diagnosis of gastric adenocarcinoma. To verify the function of RAB23 in gastric adenocarcinoma cells, MKN45 cells were transfected with shRNA-RAB23 to inhibit the expression of RAB23. Cell Counting Kit-8, transwell and flow cytometry assays were utilized to measure the cell proliferation, migration and apoptosis rate of cells, and gene and apoptosis-related protein expression levels were determined using PCR and Western blot assays.
In this study, we constructed a DEmRNAs-DEmiRNAs interaction network and identified several DEmRNA-DEmiRNA pairs involved in gastric adenocarcinoma. Based on the interaction network in this study, we identified three interactions, hsa-miR-23a-3p-PTPN4, hsa-miR-20b-5p (hsa-miR-130a-3p)-TNFRSF10B, and hsa-miR-130a-3p (hsa-miR-363-3p)-RAB23, that may be related to the pathogenesis of gastric adenocarcinoma.
PTPN4 is related to signal transduction, which regulates cell growth and differentiation, and regulates cell functions that promote apoptosis (24). The role of PTPN4 in tumors has rarely been discussed, and one study showed that it negatively regulated cell proliferation and the movement of HeLa and Hep3B cells (25). Zhu et al. reported that miR-183 exerted a proinvasive effect by inhibiting the expression of PTPN4, which may be a new therapeutic target for inhibiting the metastasis of non-small-cell lung cancer tumor stem cells (26). In our present study, the decreased expression of PTPN4 in the tissues of patients with gastric adenocarcinoma compared to normal tissues was regulated by hsa-miR-23a-3p. This finding was in line with those of other studies and emphasized the significance of PTPN4 in gastric adenocarcinoma. We speculated that PTPN4 was involved in gastric adenocarcinoma and that its role was regulated by hsa-miR-23a-3p.
It is evident that RAB23 overexpression plays a key role in malignant tumors (16). For instance, Jiang et al. (9) reported that overexpressed RAB23 increased cell growth and invasion by increasing NF-kappa B signal transduction. Jian et al. (27) also reported that RAB23 increased the migration and invasion of squamous cell carcinoma cells through the integrin 1/Rac1 pathway. More importantly, Hou et al. (22) found that RAB23 promoted tumor progression in GC. In our integrated analysis, RAB23 was significantly upregulated in tissues of patients with gastric adenocarcinoma compared to normal tissues, and this increased expression was regulated by hsa-miR-130a-3p and hsa-miR-363-3p based on the DEmiRNA-DEmRNA interaction network. Furthermore, in the GO and KEGG analyses, we found that RAB23 was enriched in the GO terms “cytoplasmic vesicle” and “intracellular vesicle”. RAB23 was related to vesicle transport, as revealed by the immunofluorescence results. More relevant to our study, another recent report confirmed that RAB23 was an upregulated gene in nonmalignant gastric tissue compared to normal gastric mucosa (28). These findings confirm that upregulated RAB23 may be related to an early stage of gastric carcinogenesis due to atrophic gastritis, which may increase the risk of GC. Importantly, Bin et al. (29) indicated that miR-367 overexpression inhibited the metastasis and invasion of gastric cancer by directly regulating RAB23. All of these data demonstrate the important role of RAB23 in the pathogenesis of gastric adenocarcinoma.
Rac1 is a member of the Rac subfamily of the Rho family. Abnormal activation of GTPases has been shown to promote cancer cell movement, invasion and metastasis in several cancers (30). Jian et al. found that Rac1 activation was crucial for Rab23-induced hepatocellular carcinoma cell migration. Upregulated Rab23 efficiently increased the active Rac1 levels, while silencing Rab23 decreased Rac1 activation. When Rac1 was silenced, the increased Hep3B cell migration induced by Rab23 expression was significantly attenuated. In squamous cell carcinoma cells, Rab23 promoted migration and invasion by regulating the integrin β1/Tiam1/Rac1 pathway (27), and Rab23 promoted migration and invasion by regulating NF-κB in bladder cancer (9). In the present study, RAB23 knockdown significantly reduced the expression level of Rac1. These findings demonstrated that Rab23 promoted gastric cancer cell migration via the PI3K-Akt signaling pathway by activating Rac1. We also speculate that Rab23 promotes the proliferation and metastasis of GC via multiple mechanisms and that Rab23 might be a useful molecular therapeutic target for gastric cancer. More studies should be performed to explore the regulation of Rab23 in gastric cancer.
Based on the GO and KEGG enrichment analyses, we found that the intracellular vesicle, vesicle-mediated transport and the PI3K-Akt signaling pathways were the most enriched pathways, and RAB23 was enriched in the GO term intracellular vesicle. Abnormal PI3K/AKT signaling can cause cancer and other diseases. The PI3K-Akt signaling pathway regulates basic cellular functions such as transcription, translation, proliferation, growth and survival. Bcl-2 and Rac1 are involved in these processes (31). As originally reported by Wu et al. (32), the PI3K/AKT signaling pathway was found to be involved in cell migration in vitro and metastasis in vivo in gastric cancer. In the GO and KEGG analyses, the PI3K-Akt signaling pathway was the most significantly enriched pathway. Our in vitro study indicated that RAB23 knockdown significantly reduced the expression of Bcl-2 and the tumor angiogenesis-promoting protein Rac1. There was no significant change in the level of PI3K protein or PI3K phosphorylation. Our immunofluorescence analysis also revealed that RAB23 knockdown affected vesicle transport. RAB23 was enriched in the GO term intracellular vesicle. The Rab protein is considered to be one of the crucial protein molecules involved in vesicle transport. Muniz et al. (33) found that in pancreatic cancer, the overexpression of RABL6A protein can promote the proliferation of tumor cells. Lin et al. (2) found that Rab25 regulated vesicle transport by changing the conformation of guanosine triphosphate, which plays an important role in the development of ovarian cancer. Jacob et al. (34) found that Rab40b was involved in the metastasis of breast cancer by regulating vesicle transport. Based on these findings, we speculated that hsa-miR-130a-3p and hsa-miR-363-3p regulate the expression of RAB23 and affect the vesicle transport-related pathways in gastric cancer cells.
The results from the metabolic analysis revealed significantly altered metabolites, including glycerol, niacinamide, nonadecanoic acid methylester and methionine. Glycerol may serve a crucial purpose in cell growth and vitality by leading to the production of ATP and lipids (35,36). Studies have shown that damage to glycerin uptake leads to loss of T cell activity, which can be reversed by glycerin administration (35). We found that RAB23 deficiency partially blocked glycerol uptake, and the decrease was associated with proliferation, injury and lipid synthesis. As mentioned earlier, there are significant differences in circulating levels of endogenous metabolites such as fatty acids, organic acids, carbohydrates, amino acids and steroids in patients with GC (37-39). These variations may indicate that the metabolism of tumor cells disrupts several metabolic pathways in patients.
In conclusion, we present evidence of the significance of RAB23 as a diagnostic marker and a therapeutic target for human gastric adenocarcinoma. The PI3K/AKT pathway is involved in promoting cell growth, inhibiting apoptosis, promoting cell migration, and carrying out RAB23-mediated metabolic changes and is important for the etiology of the disease.
Acknowledgments
Funding: The study was funded by the National Key Clinical Specialist Construction Programs of China, Fujian Provincial Natural Science Foundation Projects (2019J01452) and Fujian Provincial Natural Science Foundation Projects (2017J01287).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Lin Y, Ueda J, Kikuchi S, et al. Comparative epidemiology of gastric cancer between Japan and China. World J Gastroenterol 2011;17:4421-8. [Crossref] [PubMed]
- Park YH, Kim N. Review of atrophic gastritis and intestinal metaplasia as a premalignant lesion of gastric cancer. J Cancer Prev 2015;20:25-40. [Crossref] [PubMed]
- Ng JY, Chan DK, Tan KK. Is gastroscopy for fecal immunochemical test positive patients worthwhile? Int J Colorectal Dis 2017;32:95-8. [Crossref] [PubMed]
- Yan L, Wu K, Du F, et al. miR-384 suppressed renal cell carcinoma cell proliferation and migration through targeting RAB23. J Cell Biochem 2018. [Crossref] [PubMed]
- Zhang L, Zhang B, You W, et al. Rab23 Promotes Hepatocellular Carcinoma Cell Migration Via Rac1/TGF-β Signaling. Pathol Oncol Res 2018. [Crossref] [PubMed]
- Jiao ZH, Wang JD, Wang XJ. MicroRNA-16 suppressed the invasion and migration of osteosarcoma by directly inhibiting RAB23. Eur Rev Med Pharmacol Sci 2018;22:2598-605. [PubMed]
- Cheng L, Yang F, Zhou B, et al. RAB23, regulated by miR-92b, promotes the progression of esophageal squamous cell carcinoma. Gene 2016;595:31-8. [Crossref] [PubMed]
- Jiang Y, Han Y, Sun C, et al. Rab23 is overexpressed in human bladder cancer and promotes cancer cell proliferation and invasion. Tumour Biol 2016;37:8131-8. [Crossref] [PubMed]
- Wang Y, Wu N, Liu J, et al. FusionCancer: a database of cancer fusion genes derived from RNA-seq data. Diagn Pathol 2015;10:131. [Crossref] [PubMed]
- Stark C, Breitkreutz BJ, Reguly T, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 2006;34:D535-9. [Crossref] [PubMed]
- Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011;27:431-2. [Crossref] [PubMed]
- Lei Y, Guo P, Li X, et al. Identification of Differentially Expressed miRNAs and mRNAs in Vestibular Schwannoma by Integrated Analysis. Biomed Res Int 2019;2019:7267816. [Crossref] [PubMed]
- Liu J, Liu F, Shi Y, et al. Identification of key miRNAs and genes associated with stomach adenocarcinoma from The Cancer Genome Atlas database. FEBS Open Bio 2018;8:279-94. [Crossref] [PubMed]
- Chen C, Maimaiti A, Zhang X, et al. Knockdown of RAI14 suppresses the progression of gastric cancer. Onco Targets Ther 2018;11:6693-703. [Crossref] [PubMed]
- Zhang XY, Mu JH, Liu LY, et al. Upregulation of miR-802 suppresses gastric cancer oncogenicity via targeting RAB23 expression. Eur Rev Med Pharmacol Sci 2017;21:4071-8. [PubMed]
- Rollins J, Miskolci V. Immunofluorescence and subsequent confocal microscopy of intracellular TNF in human neutrophils. Methods Mol Biol 2014;1172:263-70. [Crossref] [PubMed]
- Zhang HH, Gu GL, Zhang XY, et al. Primary analysis and screening of microRNAs in gastric cancer side population cells. World J Gastroenterol 2015;21:3519-26. [Crossref] [PubMed]
- Song B, Yan J, Liu C, et al. Tumor Suppressor Role of miR-363-3p in Gastric Cancer. Med Sci Monit 2015;21:4074-80. [Crossref] [PubMed]
- Tsai CL, Chiu YM, Ho TY, et al. Gallic Acid Induces Apoptosis in Human Gastric Adenocarcinoma Cells. Anticancer Res 2018;38:2057-67. [PubMed]
- Tan X, Tang H, Bi J, et al. MicroRNA-222-3p associated with Helicobacter pylori targets HIPK2 to promote cell proliferation, invasion, and inhibits apoptosis in gastric cancer. J Cell Biochem 2018;119:5153-62. [Crossref] [PubMed]
- Hou Q, Wu YH, Grabsch H, et al. Integrative genomics identifies RAB23 as an invasion mediator gene in diffuse-type gastric cancer. Cancer Res 2008;68:4623-30. [Crossref] [PubMed]
- Badgwell B, Das P, Ajani J. Treatment of localized gastric and gastroesophageal adenocarcinoma: the role of accurate staging and preoperative therapy. J Hematol Oncol 2017;10:149. [Crossref] [PubMed]
- Huai W, Song H, Wang L, et al. Phosphatase PTPN4 preferentially inhibits TRIF-dependent TLR4 pathway by dephosphorylating TRAM. J Immunol 2015;194:4458-65. [Crossref] [PubMed]
- Zhou J, Wan B, Shan J, et al. PTPN4 negatively regulates CrkI in human cell lines. Cell Mol Biol Lett 2013;18:297-314. [Crossref] [PubMed]
- Zhu C, Deng X, Wu J, et al. MicroRNA-183 promotes migration and invasion of CD133(+)/CD326(+) lung adenocarcinoma initiating cells via PTPN4 inhibition. Tumour Biol 2016;37:11289-97. [Crossref] [PubMed]
- Jian Q, Miao Y, Tang L, et al. Rab23 promotes squamous cell carcinoma cell migration and invasion via integrin beta1/Rac1 pathway. Oncotarget 2016;7:5342-52. [Crossref] [PubMed]
- Evans TM, Ferguson C, Wainwright BJ, et al. Rab23, a negative regulator of hedgehog signaling, localizes to the plasma membrane and the endocytic pathway. Traffic 2003;4:869-84. [Crossref] [PubMed]
- Bin Z, Dedong H, Xiangjie F, et al. The microRNA-367 inhibits the invasion and metastasis of gastric cancer by directly repressing Rab23. Genet Test Mol Biomarkers 2015;19:69-74. [Crossref] [PubMed]
- Bid HK, Roberts RD, Manchanda PK, et al. RAC1: an emerging therapeutic option for targeting cancer angiogenesis and metastasis. Mol Cancer Ther 2013;12:1925-34. [Crossref] [PubMed]
- Pan Y, Wang N, Xia P, et al. Inhibition of Rac1 ameliorates neuronal oxidative stress damage via reducing Bcl-2/Rac1 complex formation in mitochondria through PI3K/Akt/mTOR pathway. Exp Neurol 2018;300:149-66. [Crossref] [PubMed]
- Wu X, Chen Y, Li G, et al. Her3 is associated with poor survival of gastric adenocarcinoma: Her3 promotes proliferation, survival and migration of human gastric cancer mediated by PI3K/AKT signaling pathway. Med Oncol 2014;31:903. [Crossref] [PubMed]
- Muniz VP, Askeland RW, Zhang X, et al. RABL6A Promotes Oxaliplatin Resistance in Tumor Cells and Is a New Marker of Survival for Resected Pancreatic Ductal Adenocarcinoma Patients. Genes Cancer 2013;4:273-84. [Crossref] [PubMed]
- Jacob A, Jing J, Lee J, et al. Rab40b regulates trafficking of MMP2 and MMP9 during invadopodia formation and invasion of breast cancer cells. J Cell Sci 2013;126:4647-58. [Crossref] [PubMed]
- Cui G, Staron MM, Gray SM, et al. IL-7-Induced Glycerol Transport and TAG Synthesis Promotes Memory CD8+ T Cell Longevity. Cell 2015;161:750-61. [Crossref] [PubMed]
- Hara-Chikuma M, Verkman AS. Prevention of skin tumorigenesis and impairment of epidermal cell proliferation by targeted aquaporin-3 gene disruption. Mol Cell Biol 2008;28:326-32. [Crossref] [PubMed]
- Ikeda A, Nishiumi S, Shinohara M, et al. Serum metabolomics as a novel diagnostic approach for gastrointestinal cancer. Biomed Chromatogr 2012;26:548-58. [Crossref] [PubMed]
- Song H, Peng JS, Dong-Sheng Y, et al. Serum metabolic profiling of human gastric cancer based on gas chromatography/mass spectrometry. Braz J Med Biol Res 2012;45:78-85. [Crossref] [PubMed]
- Yang T, Luo P, Li Y, et al. A serum metabolomics study of gastric cancer based on pseudotargeted liquid chromatography-mass spectrometry approach. Se Pu 2014;32:126-32. [Crossref] [PubMed]


