
Cholangiocarcinoma: anatomical location-dependent clinical, prognostic, and genetic disparities
Introduction
Cholangiocarcinoma (CCA) is a group of highly lethal epithelial malignancies that arises from complex interactions between genetic background and diverse risk factors along the biliary tree (1). Globally, CCA is a relatively rare cancer with less than <6 cases per 100,000 people in most countries, except for some Eastern countries, such as China, South Korea, and Thailand (2). To date, a number of risk factors have been identified, including hepatitis virus infection, parasitic infection, primary sclerosing cholangitis, and cholelithiasis, but approximately 70% cases occur sporadically without any risk factors (3).
Cumulating evidences indicate an increasing trend in worldwide incidence of intrahepatic CCA (iCCA), but not perihilar CCA (hiCCA) and distal CCA (dCCA) (4). In addition, therapeutic and diagnostic approaches, clinical manifestations, and prognosis of CCAs are dependent to different anatomical locations (5,6). Moreover, recent whole-exome sequencing analysis showed that mutational frequencies depended on anatomical location of CCAs (7). TP53 and KRAS alterations were significantly more frequent with worse survival rates in extrahepatic CCA, whereas IDH1, IDH2, and chromatin-remodeling gene BAP1 were typical of iCCA.
Herein, we comprehensively compared iCCA, hiCCA, and dCCA in terms of incidence, surgical resectability, therapeutic outcomes, clinicopathological features, prognostic factors, and genetic heterogeneities to support future clinical practice.
Methods
Data source
Data on 393,963 patients diagnosed with digestive system cancer between 1973 to 2015 were obtained from the Surveillance, Epidemiology, and End Results Cancer Registries (SEER) 9 Registries Program (https://seer.cancer.gov/), which is a national representative population-based database involving the following areas: San Francisco-Oakland, Connecticut, the Detroit Metropolitan Area, Hawaii, Iowa, New Mexico, the Seattle Puget Sound Area, Utah, and the Atlanta Metropolitan Area in the United States. Permission to the data was obtained in 19 Feb 2019. In addition, we also obtained genetic and clinical data of CCA from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/) (8). Institutional review board approval was not required by the Ethics Committee of Renji Hospital, considering the study design that the study did not involve any interventions. Informed consent was not applicable.
Study patient
Of the 393,963 patients with digestive cancers from the SEER database, we identified 5,626 hiCCA (Code 008, Bile Ducts Perihilar), 5,462 iCCAs (Code 007, Bile Ducts Intrahepatic), and 622 dCCAs (Code 006, Bile Ducts Distal) according to the CS Schema v0204+, which was collected under the specifications of a particular schema based on site and histology. From the TCGA database, only the patients with CCA and full follow-up information were analyzed, which include 36 iCCA, 7 hiCCA, and 2 dCCA cases.
Statistical analysis
All statistical analyses were performed using The R project for Statistical Computing (https://www.r-project.org/). All continuous and categorical variables were presented as median [Q1–Q3; interquartile range (IQR)] and n (%), respectively. Kaplan-Meier survival curves were generated for comparison of cumulative survival that was evaluated using the log-rank test. For evaluation of correlations among variables, Pearson’s correlation coefficients were adopted. Univariable and multivariable analyses were carried out using the Cox proportional hazards regression model, which were presented as hazard ratio (HR) with 95% confidence interval (CI). Relative risks were calculated along with 95% CI for identification of significant subgroups for the overall survival (OS) and tumor-specific survival (TS), defined as time from diagnosis to death due to any cause and time from diagnosis to death due to tumor-associated reasons, respectively. Significant differential expressive genes were identified in a term of log2 (fold change) >1 along with P<0.05. Prognostic networks were identified using the STRING database (https://string-db.org/) in terms of from curated databases, experimentally determined, gene neighborhood, text mining, co-expression, and protein homology (9). In addition, data regarding biological process, molecular function, cellular component, Reactome pathways, uniport keywords, PFAM protein domains, and SMART protein domains significantly associated with the prognostic networks were analyzed using the STRING database. P<0.05 was considered statistically significant.
Results
Patient characteristics
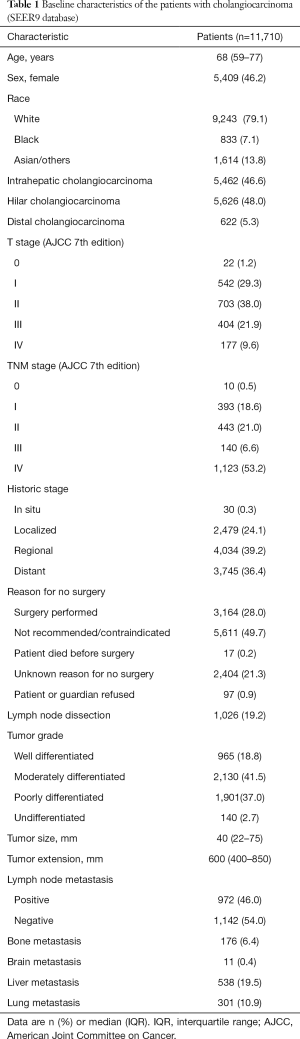
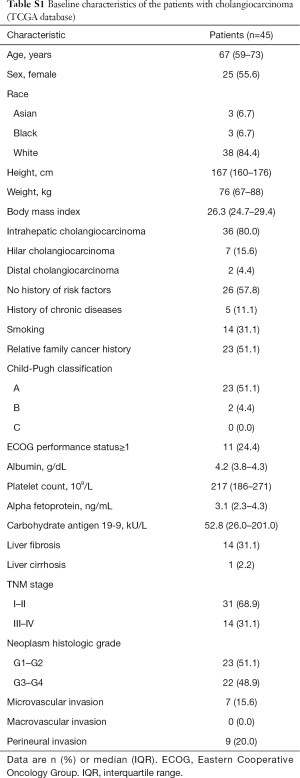
There was a total of 11,710 patients with CCA, including iCCA (n=5,462; 46.6%), hiCCA (n=5,626; 48.0%), and dCCA (n=622; 5.3%), identified from the SEER database between 1973 and 2015 with a median age of 68 years (IQR, 59–77) and 46.2% (n=5,409) of female proportion (Table 1). The proportions of White, Black, and Asian/others were 79.1% (n=9,243), 7.1% (n=833), and 13.8% (n=1,614), respectively. More than a half of the patients were diagnosed at the American Joint Committee on Cancer (AJCC) stage IV (53.2%). In addition, the most common historic stage was regional (39.2%) followed by distant (36.4%), localized (24.1%), and in situ (0.3%). The tumors were mostly moderately differentiated (41.5%) with median size of 40 mm (IQR, 22–75 mm) and extension of 600 mm (IQR, 400–850 mm). Furthermore, CCA was identified to metastasis to lymph node (46.0%), liver (19.5%), lung (10.9%), bone (6.4%), and brain (0.4%). Demographic and clinical characteristics of the patient with CCA in TCGA database that would support understanding of additional clinical and genetic analyses are shown in Table S1.
Full table
Full table
Incidence, survival rates, stage at diagnosis, and surgical outcomes
Among all CCA patients, the proportion of hiCCA (48.0%) was higher compared to iCCA (46.6%) and dCCA (5.3%; Figure 1A). Overall prognoses were relatively favorable in dCCA, moderate in hiCCA, and poor in iCCA in terms of OS and tumor-specific mortality, but short-term outcomes after surgery were highest in dCCA, followed by iCCA, and hiCCA, which revealed an accordance with the proportion of surgical candidates that only 18.5% iCCA patients received surgery (Figure 1B). The low proportion of surgery in patient with iCCA seems to be associated with relatively high proportion of distant histologic stage (43.5%) at diagnosis compared to dCCA (30.1%) and hiCCA (30.3%). For those who received surgery, 1-year OS was 67.9%, 60.4%, and 56.3% in dCCA, iCCA, and hiCCA, respectively. However, 5-year OS was highest in iCCA (16.7%) followed by hiCCA (16.4%) and dCCA (5.7%).

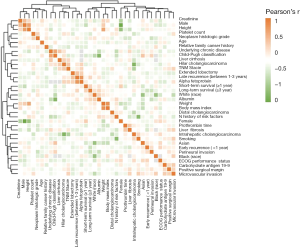
Furthermore, when ranked demographic and clinical characteristics of CCA using the Pearson’s correlation coefficient, we noticed that iCCA was in proportion to the tumor size, whereas hiCCA was in reverse proportion to the tumor size (Figure 1C). However, hiCCA revealed positive correlations with tumor extension. The correlation analysis was also performed in the TCGA database that involved factors not included in the SEER database (Figure S1). The results indicated that hiCCA is positively correlated with underlying chronic disease, Child-Pugh classification, liver cirrhosis, TNM stage, and alpha fetoprotein. In addition, positive correlations of iCCA to smoking and Eastern Cooperative Oncology Group (ECOG) performance status were observed. Furthermore, female, no history of risk factors, and TNM stage were revealed to be positively correlated with dCCA.
Anatomical location-dependent disparities in prognostic factors after surgery
In all CCA cases, age, historic stage, tumor differentiation, tumor extension, and lymph node metastasis were identified as significant prognostic factors for both OS (Figure 2) and TS (Figure S2). When stratified according to the anatomical locations, only historic stage was a significant prognostic factor for dCCA. In iCCA, age historic stage, tumor differentiation, tumor extension, and lymph node metastasis were detected. Among them, age, tumor differentiation, and lymph node metastasis were found to be independent prognostic factors for the OS. As for hiCCA, age, historic stage, lymph node dissection, tumor differentiation, tumor extension, and lymph node metastasis revealed to be significant prognostic factors.
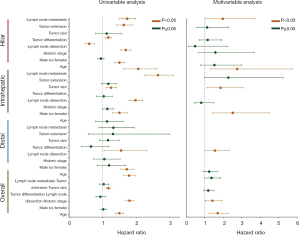
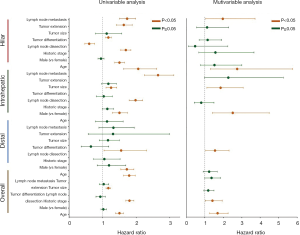
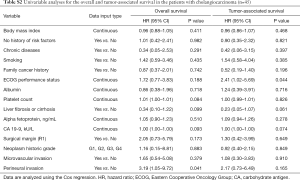
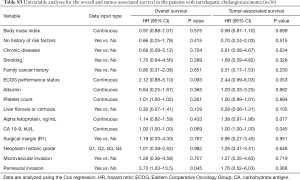
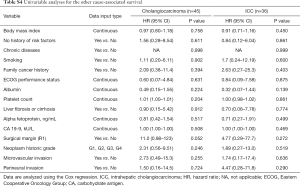
Prognostic factors for CCA were also identified in the TCGA database to confirm prognostic impact of factors not involved in the SEER database (Table S2). In the OS and RFS, perineural invasion (HR, 3.19; 95% CI, 1.05–9.72; P=0.041) and ECOG performance status (HR, 2.41; 95% CI, 1.02–5.69; P=0.044) were indicated to be significant prognostic factors, respectively. When performed the univariable analyses in iCCA only, perineural invasion was identified as a significant prognostic factor (HR, 3.73; 95% CI, 1.03–13.5; P=0.045; Table S3). Prognostic analyses were not independently performed for hiCCA and dCCA due to limited number of patients. Furthermore, we explored whether there are significant factors predictive of other cause-associated survival (Table S4). However, there were no such variable significantly predictive of other cause-associated survival.
Full table
Full table
Full table
Comparison on OS in subgroups of anatomical location-dependent CCAs
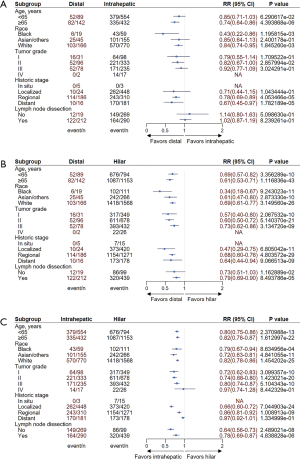
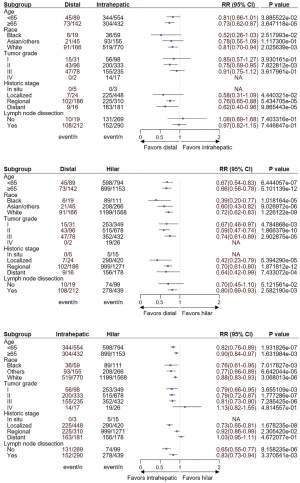
Subgroup analyses were carried out to confirm whether there are subgroups with different survival outcomes in patients with CCA who received surgery. Age ≥65 years, Black and White, and regional historic stage subgroups were identified to have significantly better OS in dCCA compared to iCCA, but the OS was comparative in other subgroups (Figure 3A). When compared dCCA to hiCCA, all subgroups demonstrated to be favorable in patients with dCCA (Figure 3B). In comparison between iCCA to hiCCA, the OS of patients with undifferentiated tumor and distant historic stage was comparative among iCCA and hiCCA, whereas iCCA was significantly favorable in the other subgroups (Figure 3C). The results for TS were also similar to OS (Figure S3).
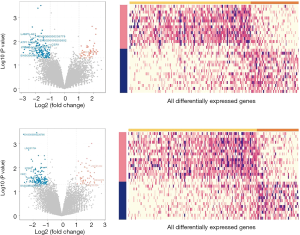
Differential expressive prognostic genes for iCCA and hiCCA
As for iCCA, we identified 179 significant prognostic genes with |log2 (fold change)| (varying from 1.04 to 2.85) and P value (varying from 0.0003 to 0.0499) for the OS (Figure S4). When screened according to recurrence of the tumor, 202 significant genes were identified. In hiCCA, 35 genes met the criteria for significant prognostic genes predictive of OS (Figure S5). Moreover, the criterion indicated 142 genes to be significant for recurrence of hiCCA. Expressions of the genes with top 10 prognostic impact for each CCA anatomical location type were highly different between iCCA and hiCCA in both for the OS that included CD24P4, KRT188P48, RIMBP2, ENPP7P10, MRPS18AP1, KCNMB2, IGLL3P, VIT, SLC18A1, and NPFFR2 for iCCA, and SLC13A1, KRT20, CA9, DLX6, PLPR1, NRSN1, RF00019, SLCO1B1, PLCXD3, and MIR122HG for hiCCA (Figure 4A) and recurrence-free survival that included CYP3A43, AL590365.1, ACSM5P1, SPATA46, RPS26P47, MIR200B, AL954705.1, RPL5P9, AL513325.1, and AC109809.1 for iCCA, and UGT1A10, GUCY1B2, MUC16, HOXC10, EVPL, NEFL, AQP5, MSLN, ASGR2, and TMEM151A for hiCCA (Figure 4B).
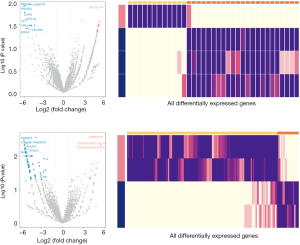

Among the identified significant genes for both OS and recurrence-free survival between iCCA and hiCCA, there was only one overlapping gene with significant impact on OS for both iCCA and hiCCA, which was PRLHR (Prolactin-releasing peptide receptor), a never studied gene in CCA (Figure 4C). In addition, SYNPR-AS1 (SYNPR antisense RNA 1), which was also never studied, was found to overlap as a significant gene with prognostic impact for recurrence of hiCCA and OS of iCCA. Furthermore, the drawn circus plots demonstrated that between-prognostic gene interactions are high in hiCCA, but low in iCCA. Collectively, these results indicate that iCCA and hiCCA have highly differential prognostic genes with different interactions.
Prognostic networks and involved processes and pathways of iCCA and hiCCA
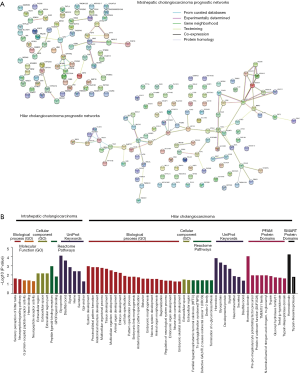
The prognostic networks for each iCCA and hiCCA were developed by adopting the identified prognostic genes in the STRING database in terms of from curative databases, experimentally determined, gene neighborhood, text mining, co-expression, and protein homology (Figure 5A). When subsequently explored significantly associated biological processes, cellular components, Reactome pathways, UniProt keywords, PFAM protein domains, and SMART protein domains, the prognostic network of iCCA was found to be significantly associated with sensory perception of bitter taste, neuropeptide signaling pathway, G protein-coupled peptide receptor activity, heme binding, neuropeptide receptor activity, extracellular region, extracellular space, extracellular region part, peptide ligand-binding receptors, GPCR ligand binding glycoprotein, disulfide bond, signal, heme, secreted, and Gpi-anchor (Figure 5B). As for the hiCCA prognostic network, system development, proximal/distal pattern formation, anatomical structure development, multicellular organismal process, tissue development, multicellular organism development, animal organ development, embryo development, cell fate specification, pattern specification process, embryonic limb morphogenesis, anterior/posterior pattern specification, behavior, embryonic morphogenesis, nervous system development, animal organ morphogenesis, regionalization, regulation of neurological system process, embryonic organ development, neuron fate commitment, embryonic skeletal system development, extracellular space, extracellular region, familial hyperphosphatemic tumoral calcinosis, Tn poly agglutination syndrome, defective GALNT12 causes colorectal cancer 1, dectin-2 family, termination of o-glycan biosynthesis, homeobox, glycoprotein, developmental protein, signal, neurotransmitter, secreted, disulfide bond, homeobox domain, pre-pro-megakaryocyte potentiating factor precursor, TMEM151 family, N0terminal barrel of ntMGAM and ctMGAM, m-glucoamyl, trypsin, glycosyl hydrolases family 31, trefoil (p-type) domain, trypsin-like peptidase domain, homeodomain, and trypsin-like serine protease. These disparities in processes, protein domains, and pathways suggest that the respective prognostic networks for iCCA and hiCCA are acted and commissioned in different ways for the recurrence of the tumor after resection and in prolongation of survival.
Discussion
CCAs are biliary cancers with cholangiocyte differentiation features (10). Among the anatomical location-dependent CCAs, iCCA is a histologically diverse malignancy that includes the conventional, intraductal, and bile ductular types (11). Unlike dCCA and hiCCA that have been reported to arise from the peribiliary glands and biliary epithelium, iCCA is considered to arise from biliary epithelial or hepatic progenitor cells (12). These different origins seem to be associated with evolving global epidemiology that the incidence of iCCA has been and is now on the rise, and this rise is suggested to be associated with risk factors of iCCA, such as hepatitis B virus infection and fatty liver diseases (13,14). Despite cumulating evidences that indicate disparities in all-round aspects of the anatomical location-dependent CCAs, direct evidences providing actual disparities are highly limited in literature due to relatively rare incidence, especially comparative studies. In a pursuit of the data, we have studied disparities in clinical characteristics, stage at diagnosis, outcomes after surgery, prognostic factors, prognostic genes, and prognostic network and its roles, functions, and pathways. We hope our study to be supportive in understanding of current status, patient selection for surgery and clinical trials, prognostic estimation, and development of new therapeutics.
Unlike hepatocellular carcinoma that the surveillance system is mature and common, CCAs are barely screened in risk populations, which derived low proportion of patients to be surgical candidates at diagnosis even in recent years (15). In the present study, the proportion of surgical candidates of dCCA, hiCCA, and iCCA remained at 37.8%, 36.3%, and 18.5%, respectively. The distant stage was present at diagnosis in 43.5% patients with iCCA, and approximately 30% in dCCA and hiCCA. Recently, Siripongsakun et al. (16) reported that surveillance by abdominal ultrasound (HR, 0.41; P=0.012) was significantly provided survival benefits, suggesting highly endemic areas for CCA to apply the surveillance system. In our point of view, the surveillance system is required not only in endemic areas, but also in all patients with known risk factors for the development of CCA, such as viral hepatitis and primary sclerosing cholangitis regarding extremely high mortality of CCA. In addition, considering rapidly increasing incidence of iCCA, the surveillance-contributed survival benefits may be magnified in terms of increase in proportion of surgical candidates.
Treatment of CCA depends on the anatomical location, but there are some surgical concerns remain further verification, including the use of lymph node dissection, liver transplantation, adjuvant chemotherapy, and target therapies (17-20). In most surgery centers, lymph node metastasis is considered a contraindication for surgical options, but it is considered resectable when it is limited to be regional in partial hospitals (21,22). In the present study, lymph node metastasis was found to be an independent prognostic factor for both OS and TS in iCCA and hiCCA, but not in dCCA. In addition, lymph node dissection significantly ameliorated survival outcomes in hiCCA only. These results suggest that either lymph node metastasis is treatable or prophylactic dissection of lymph nodes is effective in improving prognosis in patients with hiCCA. Future clinical trials are required to confirm these concerns.
Survival outcomes were significantly and relatively favorable in patients with dCCA, followed by hiCCA, and iCCA. In contrast, survival outcomes were better in iCCA compared to hiCCA when analyzed the surgical candidates only. These data are also in accordance with previous meta-analyses reporting postsurgical survival outcomes that 5-year OS was 37% and 30% for dCCA and iCCA, respectively (23,24). Therefore, there may be a disparity among anatomical location-dependent CCAs in either or both of achievement of radical resection (R0) or/and undetected metastatic tumor cells at the time of surgery, thus affecting recurrence of the tumor and OS.
Recently, the utilization of gene signatures to support treatment and prognosis prediction of various cancers has been highlighted considering its precision (25). In CCA, a three miRNA-based signature has been developed and validated to be effective in prediction of OS (26). In addition, a 30 miRNA-based signature for prediction of OS in iCCA was also introduced (27). However, we found that the prognostic network for each CCA type is highly differential, suggesting a need of independent predictive signatures for dCCA, iCCA, and hiCCA, respectively. Therefore, we have identified top genes with most effectiveness in prognostic estimation for each CCA type, which has not been described previously. We call for future clinical trials to validate the top prognostic genes and prognostic network for our data to be applied in clinical practice.
Although this study is first to comprehensively explore disparities in anatomical location-dependent CCAs, there remains some underlying limitations that need to be considered when interpreting results. First, clinical variables are not comprehensive due to retrospective nature of the study. Second, the study patients are based on the United States population. Validation is required before application of our results in patients from other regions, especially in Eastern countries. Moreover, we did not perform any functional experiments for the identified genes. The expected pathways and processes were screened from the database. Future laboratory investigations are necessary to confirm the underlying mechanisms of the top prognostic genes. Despite limitations, this study may be valuable in providing information regarding disparities in incidence, trends in disease stage at diagnosis, overall and postsurgical survival outcomes, prognostic genes, and prognostic networks and its pathways.
Conclusions
In conclusion, there are various disparities among anatomical location-dependent CCAs. These CCAs are highly differential in terms of incidence, disease stage at diagnosis, surgical resectability, susceptibility to lymph node dissection, survival outcomes, prognostic factors, and prognosis-predictive genes and its pathways. Therefore, anatomical location needs to be considered in deciding treatment strategies, predicting survival outcomes, identifying target points, and developing new therapeutic approaches for CCA.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (81570561), Top Priority Clinical Medical Center of Shanghai Health and Family Planning Commission (2017ZZ01018), Shanghai Pujiang Program (17PJD023), and Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20181806).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Institutional review board approval was not required.
References
- Labib PL, Goodchild G, Pereira SP. Molecular Pathogenesis of Cholangiocarcinoma. BMC Cancer 2019;19:185. [Crossref] [PubMed]
- Banales JM, Cardinale V, Carpino G, et al. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma. Nat Rev Gastroenterol Hepatol 2016;13:261-80. [Crossref] [PubMed]
- Khan SA, Davidson BR, Goldin RD, et al. Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut 2012;61:1657-69. [Crossref] [PubMed]
- Kirstein MM, Vogel A. Epidemiology and Risk Factors of Cholangiocarcinoma. Visc Med 2016;32:395-400. [Crossref] [PubMed]
- Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet 2014;383:2168-79. [Crossref] [PubMed]
- PDQ Adult Treatment Editorial Board. Bile Duct Cancer (Cholangiocarcinoma) Treatment (PDQ®): Health Professional Version. PDQ Cancer Information Summaries. Bethesda (MD): National Cancer Institute (US); 2002-2019.
- Valle JW, Lamarca A, Goyal L, et al. New Horizons for Precision Medicine in Biliary Tract Cancers. Cancer Discov 2017;7:943-962. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113-20. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology 2013;145:1215-29. [Crossref] [PubMed]
- Nakanuma Y, Sato Y, Harada K, et al. Pathological classification of intrahepatic cholangiocarcinoma based on a new concept. World J Hepatol 2010;2:419-27. [Crossref] [PubMed]
- Komuta M, Spee B, Vander Borght S, et al. Clinicopathological study on cholangiocellular carcinoma suggesting hepatic progenitor cell origin. Hepatology 2008;47:1544-56. [Crossref] [PubMed]
- Matsumoto K, Onoyama T, Kawata S, et al. Hepatitis B and C virus infection is a risk factor for the development of cholangiocarcinoma. Intern Med 2014;53:651-4. [Crossref] [PubMed]
- Wongjarupong N, Assavapongpaiboon B, Susantitaphong P, et al. Non-alcoholic fatty liver disease as a risk factor for cholangiocarcinoma: a systematic review and meta-analysis. BMC Gastroenterol 2017;17:149. [Crossref] [PubMed]
- Loho IM, Siregar L, Waspodo AS, et al. Current Practice of Hepatocellular Carcinoma Surveillance. Acta Med Indones 2018;50:353-60. [PubMed]
- Siripongsakun S, Vidhyarkorn S, Charuswattanakul S, et al. Ultrasound surveillance for cholangiocarcinoma in an endemic area: A prove of survival benefits. J Gastroenterol Hepatol 2018;33:1383-8. [Crossref] [PubMed]
- Lendoire JC, Gil L, Imventarza O. Intrahepatic cholangiocarcinoma surgery: the impact of lymphadenectomy. Chin Clin Oncol 2018;7:53. [Crossref] [PubMed]
- Goldaracena N, Gorgen A, Sapisochin G. Current status of liver transplantation for cholangiocarcinoma. Liver Transpl 2018;24:294-303. [Crossref] [PubMed]
- Howell M, Valle JW. The role of adjuvant chemotherapy and radiotherapy for cholangiocarcinoma. Best Pract Res Clin Gastroenterol 2015;29:333-43. [Crossref] [PubMed]
- Limpaiboon T. Epigenetic aberrations in cholangiocarcinoma: potential biomarkers and promising target for novel therapeutic strategies. Asian Pac J Cancer Prev 2012;13 Suppl:41-5. [PubMed]
- Songthamwat M, Chamadol N, Khuntikeo N, et al. Evaluating a preoperative protocol that includes magnetic resonance imaging for lymph node metastasis in the Cholangiocarcinoma Screening and Care Program (CASCAP) in Thailand. World J Surg Oncol 2017;15:176. [Crossref] [PubMed]
- Yamaoka R, Ishii T, Kawai T, et al. CD90 expression in human intrahepatic cholangiocarcinoma is associated with lymph node metastasis and poor prognosis. J Surg Oncol 2018;118:664-74. [Crossref] [PubMed]
- Zhou Y, Liu S, Wu L, et al. Survival after surgical resection of distal cholangiocarcinoma: A systematic review and meta-analysis of prognostic factors. Asian J Surg 2017;40:129-38. [Crossref] [PubMed]
- Mavros MN, Economopoulos KP, Alexiou VG, et al. Treatment and Prognosis for Patients With Intrahepatic Cholangiocarcinoma: Systematic Review and Meta-analysis. JAMA Surg 2014;149:565-74. [Crossref] [PubMed]
- Cardoso F, van’t Veer LJ, Bogaerts J, et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N Engl J Med 2016;375:717-29. [Crossref] [PubMed]
- Cao J, Sun L, Li J, et al. A novel three-miRNA signature predicts survival in cholangiocarcinoma based on RNA-Seq data. Oncol Rep 2018;40:1422-34. [PubMed]
- Zhang MY, Li SH, Huang GL, et al. Identification of a novel microRNA signature associated with intrahepatic cholangiocarcinoma (ICC) patient prognosis. BMC Cancer 2015;15:64. [Crossref] [PubMed]


