
Mesenchymal stem cells alleviate acute kidney injury via miR-107-mediated regulation of ribosomal protein S19
Introduction
The pathophysiological processes of acute kidney injury (AKI) are quite complex and consist of tubular and vascular cell damage in addition to strong inflammatory responses (1). At present, treatments of AKI primarily involve supportive treatment in the form of renal replacement therapy (2). Despite these treatments, the 5-year mortality rate of AKI patients is still more than 50%. An increasing number of experimental papers have indicated that mesenchymal stem cells (MSCs) have a good effect against AKI (3). MSCs can produce a number of immunomodulatory molecules and secrete various soluble factors that reduce apoptosis and promote mitosis. It is believed that these properties are responsible for the beneficial outcome of MSC therapy in AKI (4). MicroRNA (miRNA or miR) markers associated with repair status can illuminate some of the molecular events related to MSC protection (5). However, the characteristics of miRNA involvement in this process remain unclear.
miRNAs are posttranscriptional regulators that are crucial for the development and function of tissues and are involved in pathogenesis, including within the kidney (6). In the context of experimental and clinical AKI, many miRNAs have been associated with molecular mechanisms such as inflammation, apoptosis, angiogenesis, and fibrosis. There are several important considerations when interpreting miRNA patterns in AKI (7). To determine changes in miRNA expression in AKI, we used the miRNA prediction databases like miRanda and TargetScan to identify relevant miRNAs. From this, we hope to provide a new approach in improving our understanding of the biological contribution of miRNAs in AKI.
Transcriptome analysis and proteomics are effective tools for the systematic study of biological mechanisms. Hence, by connecting genes with protein expression, a fairly accurate functional framework of biological mechanisms can be obtained (8). Thus far, systematic analyses of transcriptional and proteomic data of AKI have been rare, but there is an urgent need to combine transcriptome and proteomic data to gain a more comprehensive understanding of biological activity in AKI. In this paper, we characterized transcriptional and proteomic changes occurring in AKI. When we combined these data, we identified those genes and proteins involved in the AKI process. These findings will help us to elucidate those miRNA and proteins responsible for cisplatin-induced AKI and can contribute to a more general understanding of miRNA function and regulatory activity on gene expression in organs or tissues of interest.
In this study, we downloaded two original databases. We chose kidney damage caused by cisplatin as a model of AKI. Changes in miRNA levels in kidney tissues from mice with cisplatin-induced AKI were compared to data from rats treated with rat bone marrow MSCs. We started a bioinformatic study to forecast the genes controlled by 44 miRNAs and adopted 5 different algorithms: miRanda, miRDB, miRWalk, RNA22, and TargetScan. We used glucose and ATP depletion as an in vitro model of AKI. After ischemic injury, differentially expressed proteins were examined in the human renal proximal tubule cell line hMSC-2. Then, we investigated the miRNA target genes and differentially expressed proteins from the two databases. Common differential gene expression (DGE) was screened by Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and the protein-protein interaction (PPI) network was adopted to ensure that the gene identified was essential.
Methods
Record sources for gene and protein analysis
Microarray data comprising significantly changed miRNAs in renal tissues from AKI mice with and without MSC treatment as reported were downloaded (9). Proteomics resources from the following cells were downloaded and compared: a human renal proximal small tube cell line (HK-2) that was subjected to chemical anoxia followed by a 3-h recovery period (anoxia → reoxygenation condition, AR), and renal cells subjected to anoxia and cocultured with human MSCs (hMSC) during the recovery period (AR + hMSC condition) (10). The identification of proteins under both conditions adopted unlabeled high definition 2D-Nano ESI-MSE.
Prediction of microRNA targets using various computational algorithms
Using 5 verified miRNA target prediction programs [miRanda-mirSVR (http://www.microrna.org/), miRDB (http://mirdb.org/miRDB/), miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/index.html), RNA22 v2 (https://cm.jefferson.edu/rna22v2.0/), and TargetScan 6.2 (http://www.targetscan.org/)], bioinformatics predictions were performed on 44 miRNA-regulated target genes.
Functional and pathway enrichment of DEG analysis
The GO and KEGG pathway analyses were derived from the Database for Annotation, Visualization and Integrated Discovery database (DAVID, http://david.abcc.ncifcrf.gov/). DAVID is capable of providing comprehensive annotations for functional and pathway interpretations. In this experiment, differentially expressed genes (DEGs) were uploaded into DAVID to carry out correlation analyses of relevant GO and KEGG pathways. The cutoff criterion was set as P<0.05.
PPI network analysis
STRING, a well-known online database for predicting PPIs (http://string.embl.de/), was used as a means for retrieving interacting genes. All candidate DEGs were entered into the STRING website, and a confidence score >0.9 or a cutoff criteria of 0.98 was set for the PPI network foundation. A simple text table of PPIs created by STRING from Cytoscape software (http://cytoscape.org/) was used to visualize the interactions.
Cell culture
HK-2 human proximal tubular cells were obtained from the American Type Culture Collection (Manassas, VA, USA). HK-2 cells were cultured in Dulbecco’s modified Eagle’s medium (supplemented with 10% FBS and antibiotics) in an atmosphere containing 5%:95% carbon dioxide/humidified air at 37 °C. For in vitro experiments, HK-2 cells were split into three groups: normal (untreated), cisplatin (7.5 mM cisplatin treatment for 6 hours), and MSC (7.5 mM cisplatin treatment and coculture with MSCs in a Transwell plate).
For the MSC cell culture, MSCs derived from the bone marrow of Sprague-Dawley (SD) rats were acquired from Cyagen Biosciences (Cyagen Biosciences, Sunnyvale, CA, USA) and cultured according to the manufacturer’s instructions. MSCs were seeded in 25-cm2 culture flasks and grown in MSC growth medium (Cyagen Biosciences) in an incubator with 5% carbon dioxide and 90% humidity at 37 °C. Experiments were performed using MSCs from passages 6–8.
Plasmid construction and luciferase assay
The RPS19 (NM_023133.2) 3'-UTRregion, including the miR-107 targeting sequence (5'- AGCAGCATTGTACAGGGCTATCA-3'), was cloned into the psiCHECKTM-2 dual-luciferase reporter plasmid (Promega, USA; cat. no. C8032) using PCR to generate psiCHECKWT-RPS19. The order 5'-TGCTGC-3' (1,103–1,109 bp) among the 3'-UTR of RPS19, which is the core binding sequence of miR-107, was replaced with CAGCAG and subcloned into psiCHECKTM-2 to create the resulting plasmid, psiCHECK-MT-RPS19. To drive site-directed mutagenesis at the miR-107 targeting site, a QuikChange® Site-Directed Mutagenesis Kit (Agilent Technologies, USA; cat. no. 201634) was used.
In the in vitro experiments, HEK293 cells were cultured in 24-well plates until they achieved approximately 80% confluence and were then transfected for 48 h with a double luciferase reporter plasmid (psiCHECK-WT-RPS19 or psiCHECK-MT-RPS19) and either miR-107 analog or miR-107 inhibitor.
The luciferase activity in the transfected cells was determined by a Dual-Luciferase Reporter Assay System (Promega cat. no. E1910), and Renilla luciferase activity was used to normalize the firefly luciferase activity.
Small RNA transfection
To interfere with miR-107 activity, HK-2 cells at 50% confluence were transfected with miR-107 antagonists (10 nM) or scrambled oligonucleotides (GenePharma, Shanghai, China) using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions.
Rat model of AKI
Six- to eight-week-old male SD rats were obtained from the Beijing Vital River Laboratory Animal Technology Co., Ltd. and provided water and food ad libitum. All animal experiments were accepted by the Animal Ethics Committee of the People’s Hospital of Zhengzhou University, (6–8 rats per group). Cisplatin (Sigma-Aldrich) was injected intraperitoneally into rats at a dose of 6 mg/kg to induce AKI. In the MSC group, MSCs were injected via the tail vein 1 day after cisplatin treatment. All the animals were dead on the 3rd day after cisplatin treatment.
Western blotting
The homogenate was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis through a 10% gel, and the proteins were transferred to nitrocellulose membranes. Then, the membranes were incubated with 5% skim milk for 1 h at room temperature. Anti-Bcl-2-associated X (Burlington) and anti-Bcl-2, anti-proliferating cell nuclear antigen (PCNA) (Abcam, Cambridge, UK) were incubated at 4 °C. The bands were observed with enhanced chemiluminescence reagent after treatment with peroxidase-coupled secondary antibody.
Measurement of rat renal function
Serum creatinine (Scr) and blood urea nitrogen (BUN) levels were measured by commercial kits (sarcosine oxidase-peroxidase-antiperoxidase; Zixing, Shanghai, China) according to the manufacturer’s instructions.
PAS staining and histology scoring
Renal tissues were soaked in 10% neutral formalin and embedded in paraffin. Periodic acid-Schiff (PAS) staining for 4-µm thick renal sections was analyzed, and 10 randomly selected non-overlapping fields (200×) in the cortex and medulla were viewed. After that, the degree of injury (loss of brush border, tubular expansion, mold formation, cell lysis) was scored as follows: 1≤25%, 2=25–50%, 3=50–75%, 4≥75%.
Statistical analysis
The results are presented as mean value ± standard deviation. IBM SPSS Statistics 17.0.2 software (IBM Corporation, Armonk, NY, USA) was used for analysis. The parameter data were compared several times with one-way ANOVA. The analyses were considered statistically significant when P<0.05.
Results
Identification of microRNAs in cisplatin-induced AKI rats and the predicted target genes
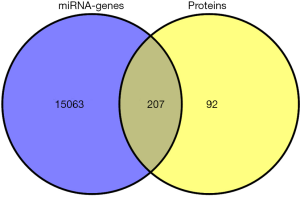
Kidney tissues from the cured AKI rats with and without MSC treatment showed differential expression among 44 miRNAs (36 increased, 8 decreased). These miRNAs were then entered into the miRanda, miRDB, miRWalk, RNA22, and TargetScan databases to predict the genes they regulate, and 15,270 target genes were identified.
Identification of variously expressed proteins in HK-2 cells treated with AR + hMSC
HK-2 cells cultured in the absence of glucose and ATP were the in vitro model of AKI. After analysis of unlabeled high definition 2D-Nano ESI-MSE protein, 310 differentially expressed proteins were recognized in HK-2 cells treated with hMSC after injury. Among them, 262 were more highly expressed, and 48 showed lower expression after treatment with AR + hMSC.
Following a careful analysis of the 15,270 predicted microRNA target genes and the 310 differentially expressed proteins, we ultimately screened 207 common elements (DEGs) after multiple testing corrections. After DAVID proofreading, 301 proteins corresponded to 299 genes. The 207 DEGs became our key focus in this study (Figure 1).
miRNAs regulate gene expression by binding to mRNAs and either directly degrading the mRNA transcript or inhibiting protein translation. We reanalyzed the 44 identified microRNAs and 207 DGEs with the miRNAs and their target genes. However, we recognized that the increase in the levels of 28 miRNAs was related to the downregulation of 30 target DGEs, while the downregulation of 6 miRNAs was associated with the upregulation of 125 DGEs.
Gene Ontology and signaling pathway enrichment analysis of DEGs
Using GO enrichment analysis, DAVID returned abundant information regarding the molecular functions, biological processes, and cellular components of 155 DGEs. GO enrichment analysis revealed that a number of the DEGs were critically concentrated in extracellular vesicles, extracellular organelles, extracellular exosomes, vesicles, membrane-bound vesicles, extracellular regions, cytosol, RNA binding, poly(A) RNA binding, and extracellular regions (Figure 2A). As shown in Figure 2B, in the biological process analysis, DEGs were primarily concentrated on cellular function; the regulation of biological processes, metabolic processes, and macromolecule metabolism; and reactions to stimuli. In the molecular function analysis, DEGs were primarily concentrated in protein binding, nucleic acid binding, hydrolase activity, catalytic activity, and structural molecule activity. In the cell component group, DEGs were primarily concentrated in the cell, cytoplasm, intracellular region, nucleus, membrane, and extracellular region.
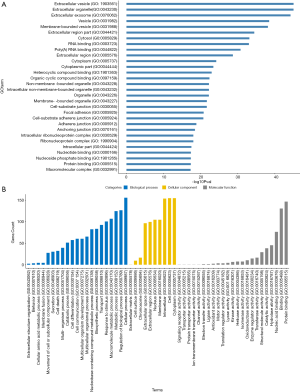
The concentration of DEG function and signal pathways were further assessed via KEGG pathway analysis. As shown in Figure 3, a large number of genes were primarily concentrated in oocyte meiosis, protein procedure in endoplasmic reticuli, pathogenic Escherichia coli infection, ribosome, salmonella infection, regulation of actin cytoskeleton, hippo signaling pathway, shigellosis, adrenergic signaling in cardiomyocytes and proteoglycans in cancer. The downregulated genes were mainly enriched in RNA transport, ribosome, spliceosome, cysteine and methionine metabolism, pathogenic Escherichia coli infection, biosynthesis of amino acids, gap junction, vitamin B6 metabolism, apoptosis, and phagosome.

PPI network construction
The PPI network is established by adopting STRING to perform a systematic analysis of DEG function. The degree of the proteins in the PPI network as a node indicates the number of proteins interacting with a particular node. The hub node is regarded as having a high node degree. Using a combined score truncation >0.9, the PPI network of DEGs was established and consisted of 105 nodes and 357 edges. The 105 nodes comprise a total of 92 genes with increased expression and 13 genes with decreased expression (Figure 4A). A total of 27 central node genes were screened out by screening >10 degrees in 105 nodes, and they were EIF4A3, HSP90AA1, RPL8, RPS23, RPL10, RPL27A, ACTB, RPL13A, RPL13,RPS14, RPL22, RPL31, RPL37A, EIF4A2, EFTUD2, YWHAE, YWHAG, EIF3B, EIF3A, EIF2S3, MAPK1, TUBA1A, EEF2, EEF1A1, TUBA4A, PRKACA and SNRPE. We found that RPL8, RPS23, RPL10, RPL27A, RPL13A, RPL13, RPS14, RPL22, RPL31, and RPL37A, all of which are members of the ribosomal protein family.
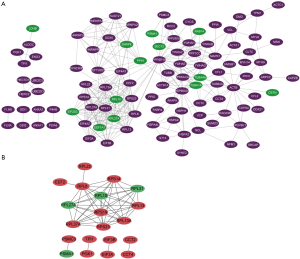
Another PPI network with 20 nodes and 51 edges was constructed using the cutoff combinatorial score >0.98. Of the 20 nodes, 27 central node genes were screened according to the >9 degree criteria, and they were RPL8, RPL10, RPL13, RPL13A, RPL27A, RPL31, RPL37A, RPS14, RPS19, and RPS23 (Figure 4B), all of which are ribosomal proteins.
These STRING results indicate that ribosomal proteins might have some important functions in the process of AKI. The pathway analysis shows that the ribosome pathway is significantly altered in AKI. According to these results, ribosomal proteins may be an important research target.
RPS19 is a larval purpose of miR-107 during AKI
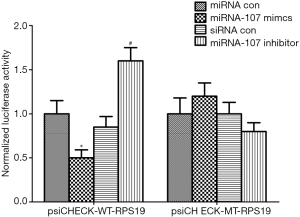
Based on the literature, we concentrated on ribosomal proteins and chose RPS19 for further study. According to the miRNA target prediction program TargetScan, RPS19 was an alternative target gene for miR-107. Thus, we verified the direct interaction between RPS19 and miR-107. The 3'-UTR of RPS19 (predicted to interact with miR-107) was cloned into a luciferase reporter vector and co-transfected with miR-107 antagonist or a scramble oligonucleotide into HEK293 cells. Inhibiting miR-107 expression in HEK293 cells improved the luciferase expression level from the plasmid containing the RPS19 3'-UTR. In addition, there was no significant difference in luciferase expression between RPS19 3’-UTR and mutant cells (Figure 5). These findings suggest that miR-107 can interact finely with RPS19 mRNA and inhibit the expression of RPS19 at the posttranscriptional level.
miR-107 is downregulated, and RPS19 is upregulated in cisplatin-induced AKI rats treated with MSCs
We induced a cisplatin-induced AKI rat model. No significant tubular injury was found in the control group, but tubule necrosis, tubule dilatation, and cast formation were observed in cisplatin-induced AKI rats. Significant improvement in renal tubular injury was observed in cisplatin-treated rats administered with MSCs (Figure 6A). The levels of serum creatinine (Scr) and BUN in AKI rats were markedly higher than those in the control group. MSC transplantation significantly decreased serum Cr and BUN levels in AKI rats (Figure 6B,C).
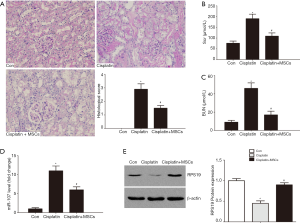
Using quantitative RT-PCR, we confirmed the increase in miR-107 levels in the kidneys of AKI rats and contrasted them with those in the control rats. We also demonstrated that miR-107 expression was decreased in the kidneys of rats treated with bone marrow MSCs (Figure 6D).
Moreover, we detected the protein expression of RPS19. Western blot analysis demonstrated that the protein levels of RPS19 were decreased in the kidneys of cisplatin-induced AKI rats but were increased after MSC treatment (Figure 6E).
miR-107 is downregulated and RPS19 is upregulated in HK-2 cells cocultured with MSCs
To confirm the role of MSCs in vitro, we treated HK-2 cells with cisplatin. miR-107 expression in HK-2 cells was consistent with that observed in vivo: it increased after cisplatin was added and was downregulated when the cells were cocultured with MSCs (Figure7A). We found that the expression level of RPS19 protein in the cisplatin group was higher than that in the control group, and the presence of cocultured MSCs increased RPS19 expression in the HK-2 cells. The levels of PCNA in the cisplatin group fell; however, they returned to normal when MSCs were included in the culture. In contrast, the Bax/bcl-2 ratio was upregulated in the cisplatin group but downregulated in the MSC group (Figure 7B), indicating that cisplatin could decrease the effects of miR-107, reduce RPS19 levels, and increase apoptosis, all of which were reversed by MSC intervention.
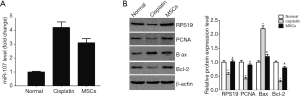
miR-107 regulates RPS19, thus affecting cell apoptosis
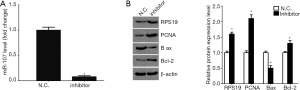
To investigate the possible role of miR-107 in cisplatin-induced AKI, we adopted miR-107 antisense oligonucleotides and assessed the regulatory role of miR-107 on RPS19 in HK-2 cells. We used real-time RT-PCR to verify the effectiveness of the miR-107 inhibitor in HK-2 cells by using the scramble fragment (NC) as the control (Figure 8A). We also detected RPS19 protein expression after miR-107 inhibition under cisplatin treatment. The level of RPS19 in the cisplatin group was higher than that in the control group. After inhibiting miR-107 expression, the expression of RPS19 was increased in the cisplatin-treated group. Moreover, we detected the expression of the proliferation-related protein, PCNA, and apoptosis-related proteins, Bax and Bcl-2. Inhibition of miR-107 improved PCNA expression and decreased the Bax-Bcl-2 ratio. These findings show that miR-107 may target RPS19 in response to potential apoptosis in cisplatin-injured HK-2 cells (Figure 8B).
Discussion
AKI leads to serious renal damage, and MSCs have improved outcomes in AKI models (11). However, the mechanism of action of bone marrow-derived MSCs in the repair of renal injury remains to be further studied (12). In this paper, we combined the transcriptome and proteomics data to analyze AKI models. Renal tissues were collected from AKI rats treated with or without MSCs and were then analyzed using gene microarray analysis to identify miRNAs. miRNA target genes were predicted by miRNA target prediction programs. Proteomic data of differentially expressed proteins were identified in an in vitro model of MSC-treated AKI. Using AKI proteomics, transcriptomics, and miRNA data, we could thoroughly analyze the relationships among these genes or proteins and discuss a few of the mechanisms by which these genes or proteins contribute to AKI.
Because proteomic and transcriptomic data can provide expression patterns of a number of genes and proteins within the human genome simultaneously, we connected transcriptome data with the study of Zhu et al. (9) along with proteomic data from da Costa et al. (10) to evaluate gene or protein levels after MSC therapy in AKI. Our study found 207 DEGs between the AKI model and MSC-treated models after data processing. Concentration analysis of GO indicated that a number of DEGs were concentrated in the following locations: extracellular vesicles, extracellular organelles, extracellular exosomes, vesicles, membrane-bound vesicles, extracellular regions, cytosol, RNA binding sites, and poly(A) RNA binding sites. According to the KEGG pathway analysis, we found that the ribosome pathway may be important in the process of MSC-treated AKI. In addition, we identified the ribosomal proteins RPL8, RPS23, RPL10, RPL27A, RPL13A, RPL13, RPS14, RPL22, RPL31, RPL27A, RPL31, RPL37A, RPS14, RPS19, and RPS23 in the PPI network constructed with STRING. On the basis of many reported studies, RPS19 may be an important ribosomal protein in the process of AKI. According to the miRNA target prediction program, RPS19 was a candidate target gene of miR-107. The findings show that these genes and pathways may play significant roles in the development of AKI.
miRNAs are a family of small noncoding single-stranded RNAs that negatively regulate protein expression by targeting mRNA transcripts and either inhibit translation or degrade the RNA transcript (13). Reports show that miRNAs play a significant role in the progression of renal disease (14). Wang et al. reported an increased level of miR-107 in circulating endothelial cells in patients with sepsis-related AKI (15). Hsieh et al. reported elevated whole blood miR-107 level in mice exposed to LPS (16). We also identified stable changes in miR-107 by real-time RT-PCR analysis of both an AKI rat model and a cell culture model. The expression of miR-107 in the kidney tissue of AKI slowly increased but fell in rats treated with MSCs. There are many potential functions for any given miRNA. A method for predicting miRNA function helps to narrow down the possible targets for experimental verification, which is key in recognizing miRNA target interactions (17). In our thorough investigation of the possible target proteins of miR-107, we found, via TargetScan analysis, that RPS19 is one of its primary target proteins.
From microorganisms to mammals, ribosomal proteins have a variety of functions in vitro, including chromatin structuring, gene transcription, RNA processing and splicing, and posttranslational modification (18). RPS19 is a 16 kDa protein that is a well-known constituent of the 40S ribosomal subunit (19). Lv et al. confirmed that renewable RPS19 behaves as a kind of MIF inhibitor, which can block MIF signal transduction and inhibit the infiltration and activation of macrophages and T cells, causing the inhibition of progressive renal injury induced by glomerular mesangium and basement membrane (GBM) semiluminal glomerulonephritis (GN) (20). Studies such as those conducted by Li have suggested that rats treated with RPS19 have renal insufficiency and weakened pathological changes when subjected to cisplatin-induced AKI (21). Based on the predicted genes from TargetScan, we hypothesized RPS19 to be a target gene of miR-107. We tested this hypothesis with a dual luciferase reporter assay and confirmed that miR-107 directly controls RPS19. We also hypothesized that MSCs inhibit apoptosis mediated by miR-107, which itself regulates RPS19. We found that miR-107-regulated expression of RPS19 is a possible mechanism of cisplatin-induced AKI and that inhibiting miR-107 is the possible mechanism by which MSCs ameliorate the effects of cisplatin-induced AKI. In this study, downregulation of miR-107 increased the level of RPS19, which inhibited apoptosis-related proteins in the presence of cisplatin. The present research indicates that MSCs can decrease miR-107 expression by promoting cell proliferation and by directly regulating RPS19 in cisplatin-induced AKI.
The observations from our study provide insights into the construction of miRNAs in MSC-mediated AKI repair and suggest that miR-107 and RPS19 play a significant role in cisplatin-induced AKI. Therefore, miR-107-regulated expression of RPS19 may represent a possible therapeutic approach for AKI in the future.
Acknowledgments
Funding: This study was supported by the Joint Provincial and Ministerial Project of China (No. SBGJ2018062).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work, and the questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments were accepted by the Animal Ethics Committee of the People’s Hospital of Zhengzhou University.
References
- Schrezenmeier EV, Barasch J, Budde K, et al. Biomarkers in acute kidney injury - pathophysiological basis and clinical performance. Acta Physiol (Oxf) 2017;219:554-72. [Crossref] [PubMed]
- Marengo M, Dellepiane S, Cantaluppi V. Extracorporeal Treatments in Patients with Acute Kidney Injury and Sepsis. Contrib Nephrol 2017;190:1-18. [Crossref] [PubMed]
- Wang Y, He J, Pei X, et al. Systematic review and meta-analysis of mesenchymal stem/stromal cells therapy for impaired renal function in small animal models. Nephrology (Carlton) 2013;18:201-8. [Crossref]
- Lee HK, Lim SH, Chung IS, et al. Preclinical efficacy and mechanisms of mesenchymal stem cells in animal models of autoimmune diseases. Immune Netw 2014;14:81-8. [Crossref]
- Ferguson SW, Wang J, Lee CJ, et al. The microRNA regulatory landscape of MSC-derived exosomes: a systems view. Sci Rep 2018;8:1419. [Crossref] [PubMed]
- Bhatt K, Mi QS, Dong Z. microRNAs in kidneys: biogenesis, regulation, and pathophysiological roles. Am J Physiol Renal Physiol 2011;300:F602-10. [Crossref]
- Popović M, de Marco A. Canonical and selective approaches in exosome purification and their implications for diagnostic accuracy. Transl Cancer Res 2018;7:S209-25. [Crossref]
- Garcia-Seco D, Chiapello M, Bracale M, et al. Transcriptome and proteome analysis reveal new insight into proximal and distal responses of wheat to foliar infection by Xanthomonas translucens. Sci Rep 2017;7:10157. [Crossref] [PubMed]
- Zhu Y, Yu J, Yin L, et al. MicroRNA-146b, a Sensitive Indicator of Mesenchymal Stem Cell Repair of Acute Renal Injury. Stem Cells Transl Med 2016;5:1406-15. [Crossref] [PubMed]
- da Costa MR, Pizzatti L, Lindoso RS, et al. Mechanisms of kidney repair by human mesenchymal stromal cells after ischemia: a comprehensive view using label-free MS(E). Proteomics 2014;14:1480-93. [Crossref] [PubMed]
- Togel FE, Westenfelder C. Mesenchymal stem cells: a new therapeutic tool for AKI. Nat Rev Nephrol 2010;6:179-83. [Crossref] [PubMed]
- Guillamat-Prats R, Camprubí-Rimblas M, Bringué J, et al. Cell therapy for the treatment of sepsis and acute respiratory distress syndrome. Ann Transl Med 2017;5:446. [Crossref] [PubMed]
- Ge QM, Huang CM, Zhu XY, et al. Differentially expressed miRNAs in sepsis-induced acute kidney injury target oxidative stress and mitochondrial dysfunction pathways. PLoS One 2017;12:e0173292. [Crossref] [PubMed]
- Godwin JG, Xupeng G, Kristin S, et al. Identification of a microRNA signature of renal ischemia reperfusion injury. Proceedings of the National Academy of Sciences of the United States of America 2010;107:14339-44. [Crossref] [PubMed]
- Wang S, Zhang Z, Wang J, et al. MiR-107 induces TNF-α secretion in endothelial cells causing tubular cell injury in patients with septic acute kidney injury. Biochem Biophys Res Commun 2017;483:45-51. [Crossref] [PubMed]
- Hsieh CH, Rau CS, Jeng JC, et al. Whole blood-derived microRNA signatures in mice exposed to lipopolysaccharides. Journal of Biomedical Science 2012;19:69. [Crossref] [PubMed]
- He S, Zeng S, Zhou ZW, et al. Hsa-microRNA-181a is a regulator of a number of cancer genes and a biomarker for endometrial carcinoma in patients: a bioinformatic and clinical study and the therapeutic implication. Drug Des Devel Ther 2015;9:1103-75. [PubMed]
- Bhavsar RB, Makley LN, Tsonis PA. The other lives of ribosomal proteins. Human Genomics 2010;4:327-44. [Crossref] [PubMed]
- Khan A, Ali A, Junaid M, et al. Identification of novel drug targets for diamond-blackfan anemia based on RPS19 gene mutation using protein-protein interaction network. BMC Syst Biol 2018;12:39. [Crossref] [PubMed]
- Lv J, Huang XR, Klug J, et al. Ribosomal protein S19 is a novel therapeutic agent in inflammatory kidney disease. Clin Sci (Lond) 2013;124:627-37. [Crossref] [PubMed]
- Li J, Tang Y, Tang PMK, et al. Blocking Macrophage Migration Inhibitory Factor Protects Against Cisplatin-Induced Acute Kidney Injury in Mice. Mol Ther 2018;26:2523-32. [Crossref] [PubMed]


