
A proposed synergistic effect of CSF1R and NMUR2 variants contributes to binge eating in hereditary diffuse leukoencephalopathy with spheroids
Introduction
Binge eating (BE) is the most common type of eating disorder and is characterized by a loss of control over eating, during which the individual consumes an unusually large amount of food within a short period of time in the absence of regular inappropriate compensatory behaviors (1). BE is seldomly mentioned among existing disease entities but is associated with substantial psychiatric symptoms (2,3). A strong genetic contribution has been suggested for BE (4,5). Family and twin studies have provided evidence that BE aggregates strongly in families independently of obesity (6). Variations in the melanocortin-4 receptor gene (MC4R) (7) and several other genes, including TPH-1, HDAC4, ESRRA, DRD2, and BDNF, and the serotonin-transporter-linked polymorphic region (5-HTTLPR) of SLC6A4 (1,8), have been suggested to be associated with eating disorders, including BE (9). There are also genes that are functionally related to BE that were not screened in BE patients, such as neuromedin U receptor 2 (NMUR2) (10).
Hereditary diffuse leukoencephalopathy with spheroids (HDLS) is an autosomal dominant white matter disease caused by mutations in the protein tyrosine kinase domain of the colony-stimulating factor 1 receptor (CSF1R), encoded by the CSF1R gene on chromosome 5q32.4. HDLS is characterized by behavioral changes, memory decline, executive dysfunction, depression, motor impairments, and seizures (11,12). BE is not a common manifestation in HDLS. In a review study summarizing 51 HDLS patients (13), seven patients had documented hyperorality and dietary changes that are viewed as a part of the behavioral variant of frontotemporal dementia (bvFTD) features. HDLS is representative of primary microgliopathies, and patients’ cranial MRI findings are consistent with frontal-predominant leukoencephalopathy and concomitant cerebral atrophy (14).
As BE disorder has always been complicated by psychiatric comorbidities, single-gene disease models with a BE phenotype would be realistic for studying its genetic mechanism with the benefit of a pure hereditary and clinical background. Recently, we encountered a family with CSF1R-related leukoencephalopathy that manifested uniformly with BE-onset dementia. In this study, we investigated the genetic factors associated with this rare phenotype.
Methods
We obtained informed and written consent from available family members, and the study was approved by the Ethical Review Board of Peking Union Medical College Hospital. Detailed clinical manifestation of all the affected family members had been recorded. Genomic DNA was extracted from peripheral blood leucocytes using a QIAamp DNA Blood Midi Kit (Qiagen, Valencia, CA, USA) according to standard protocols. Exome enrichment of 3 µg genomic DNA (individual II-4, III-3, III-4 and III-5) was performed using the Agilent SureSelect Human Exon Kit capture library. The capture library was then loaded onto the Hiseq2000 platform. Raw image files were processed by Illumina base calling Software 1.7. Reads were aligned to a human reference sequence (UCSC assembly hg19, NCBI build 37), and genotypes were called using SAMtools or GATK. Variants were annotated using SeattleSeq (http://sngs.washington.edu/SeattleSeqAnnotation131/), SIFT (http://sift.jcvi.org/), and PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/). Population frequencies for each variant were determined in dbSNP, the 1000 Genomes Project, and the NHLBI Exome Sequencing Project version ESP6500 exome variant server [http://evs.gs.washington.edu/EVS/ (1 Dec 2013)]. For each sample, the coverage of the targeted region was more than 99.1%. Over 58% of at least 83 million reads were uniquely mapped to the targeted human exon regions to give an average depth of 70. Under such coverage, approximately 97% of targeted regions were covered by 4 reads or more, and 91% were covered by more than 20 reads.
Results
Clinical presentations
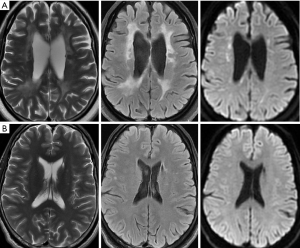
The family demonstrated an autosomal dominant inheritance pattern (Figure 1). The proband was a 43-year-old female who became apathetic and less social for two years. She often sat alone at family gatherings and showed diminished interrelatedness. With disease progression, cognitive impairment and urinary incontinence ensued. Her short-term memory declined quickly, and she was incapable of doing housework such as cooking meals. The patient gained weight, over 10 kg within 3 months, before her visit to our hospital because of compulsive eating at every mealtime. Her family also reported that she experienced episodes of unconsciousness with suspected convulsion. Neuropsychological testing revealed depression and comprehensive cognitive decline, including marked deficits in attention and executive dysfunction, short- and long-term memory decline, nonfluent aphasia, dyscalculia and a loss of visual space, with an MMSE score of 15/30 and an MoCA score of 13/30. Other neurological examinations were essentially unremarkable, with no paralysis, ataxia or sensory deficits. Routine blood and urine laboratory examinations were normal. Electroencephalography showed diffused slowing waves. Cranial MR imaging (Figure 2A) showed severe symmetric confluent bifrontal and biparietal white matter lesions, with marked cortical and subcortical atrophy. The corpus callosum was also involved. There were several foci showing hyperintensities on T2-weighted and fluid-attenuated inversion recovery (FLAIR) imaging as well as diffusion-weighted imaging with a decreased apparent diffusion coefficient. No contrast enhancement was seen. Subsequently, the activity of enzymes in the serum and leukocytes, including α- and β-galactosidase, α- and β-glucosidase, α-fucosidase, α-mannosidase, β-hexosaminidase, arylsulfatase A and galactocerebroside, as well as the serum level of very-long-chain fatty acids, were tested and were all within normal ranges. HDLS was diagnosed based on her clinical and genetic findings. The patient was given topiramate and antidepression treatment.
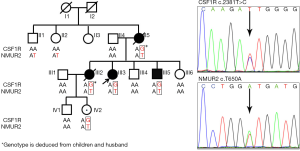
Her older sister (family member III-1) showed similar symptoms at the age of 37 and was admitted to our hospital in 2006. Personality changes and cognitive impairment were the initial features. She was described as being apathetic, having difficulties at work, and losing personal hygiene. She gained more than 10 kg of weight within 10 months because of compulsive eating. Brain MRI showed significant patchy and confluent bifrontal and biparietal periventricular white matter lesions, with moderate cortical and subcortical atrophy. Lumbar puncture and stereotactic brain biopsy of the left frontal lobe were performed during hospitalization. CSF examination was unremarkable. Microscopic examination showed a reduction in and vacuolar degeneration of cortical neurons and reactive astrocytosis with axonal dilations (spheroids) in the subcortical white matter. The spheroids were GFAP positive but were negative for ubiquitin. No infarction, demyelination or lymphocytic infiltrates were observed. The cortical and medullary vessels were unremarkable. Pathological findings were in line with sclerosis leukoencephalopathy. Within five years after symptom onset, she became wheelchair-bound, had severe dementia with communication difficulties, and suffered from frequent seizure attacks before dying.
The proband’s mother showed the exact similar pattern of disease progression. She began to show symptoms at the age of 58. After going through all the stages of binge eating, dementia and epilepsy, she finally became unresponsive and died five years after symptom onset.
The proband’s 39-year-old younger brother reported no symptoms other than transient numbness in the right limbs. His brain MRI (Figure 2B) showed asymmetric and patchy lesions in the frontal periventricular white matter, which were identified as presymptomatic changes of HDLS (14). He was thus diagnosed with presymptomatic HDLS.
Genetic analysis
Whole exome sequencing (WES) data were estimated by strict screening in which only predicted pathogenic rare nonsynonymous, splicing and frameshift variants were selected. Potentially pathogenic variants in five genes (CSF1R, EML1, NMUR2, PITPNM2 and SPDYC) were filtered out by exome sequencing in the four family members. Further analysis of linkage disequilibrium in the family by Sanger sequencing of the DNA of six family members (II-1, II-2, III-1, III-6, IV-1, IV-2) suggested a recurrent mutation in CSF1R, c.2381T>C(p. I794T) (15), cosegregated with the disease. The family was therefore genetically diagnosed with HDLS. Notably, individual IV-2, a 28-year-old female who is currently pregnant, is an obligate carrier of CSF1R mutation with no symptoms yet.
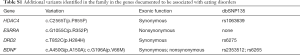
When inspecting rare variants that may be associated with BE, one single nucleotide variation (SNV), c.T650A(p. I217N), in NMUR2, was identified as being transmitted from the proband’s maternal line. The reported frequency is 0.046% (1/2,178) in dbSNP135 and 0.05% in the 1000 Genome Project database, which made it potentially pathogenic. We further examined the SNV in a noncommercial in-house database composed of Chinese Han individuals without neurological disorders and obtained a variant frequency of 0.3% (9/3,004). Single-nucleotide polymorphisms (SNPs) in the genes documented to be associated with eating disorders, including MC4R, TPH-1, HDAC4, ESRRA, DRD2, BDNF and SLC6A4, were also inspected for the proband. SNPs in those genes are listed in Table S1. However, these SNPs were either inherited from the proband’s paternal line so that they were not involved with the disease or lost in the affected individuals (as in the genes of MC4R and SLC6A4). Since the disease phenotype mimics bvFTD, we also examined the hexanucleotide GGGGCC repeat expansion in C9ORF72 by repeat-primed PCR with a previously reported method (16). The result was negative with a 6-time repeat expansion size. The WES data of three genetically confirmed HDLS patients without BE manifestations were also investigated for NMUR2 rare variants. They had 5, 2 and 1 SNPs, respectively. No rare variants with a minor allele frequency (MAF) less than 0.5% documented in the ExAC or dbSNP databases were identified.
Full table
Conclusions
Neuromedin U (NMU) is a gut-brain peptide that regulates food intake (17) and body weight (18) via the peripherally expressed NMU receptor 1 (NMUR1) and the central NMUR2. NMUR2 knockdown rats demonstrated significantly greater binge-type food consumption of the high-fat diet and showed a greater preference for higher-fat food than controls (10). However, despite its functional involvement with eating abnormalities, the NMUR2 gene has never been screened in patients with associated disorders. This was mostly because BE in most scenarios is complicated with psychiatric comorbidities, which would infer a polygenic or additive genetic pattern of inheritance. Adults with BE usually experience anxiety disorders, mood disorders, impulse control disorders, or substance use disorders. These complex phenotypic backgrounds made genetic analysis impossible for BE studies.
In the family that we studied, HDLS patients had BE-onset dementia as a uniform presentation, and had a similar pattern of disease progression. A potentially pathogenic variation in NMUR2 was carried by the first- and second-degree relatives of the proband’s mother along with the CSF1R mutation. No other potentially pathogenic variants in the existing BE-associated genes were detected to cosegregate in the family. Additional analysis of three HDLS patients without BE symptoms revealed no rare variants in NMUR2. We therefore propose that these overlapping phenotypes of HDLS dementia and BE may be explained by shared genetic risk factors and that NMUR2 is a genetic modifier of HDLS caused by CSF1R mutation. Carrying the very rare NMUR2 variant would modify the disease phenotype, in the current case, contributing to the manifestation of BE in HDLS.
It should be noted that family members with the NMUR2 variant but without the CSF1R mutation did not present with BE, suggesting that having NMUR2 rare variant(s) would not be sufficient for developing the phenotype. In HDLS, diffused white matter lesions with frontal and parietal lobe predominance involving the periventricular deep white matter and the dilation of the lateral ventricles detected by brain imaging were confirmed (19). Neuropathological findings include prominent lesions with myelin and axonal damage, neuroaxonal swelling (spheroid formation), and pigmented macrophages and glia (20). On cerebral MRI scans, the white matter lesions without gadolinium enhancement can be scattered and asymmetric during the initial stages of the disease but later become confluent, diffuse, and more symmetric. A thinning of the corpus callosum with signal abnormalities is observed even in the early stages of the disease (21). These are indications for diffuse disruption of the structure and function of the brain in HDLS. NMUR2 is a high-affinity receptor for NMU found in discrete regions of the central nervous system, particularly the paraventricular nucleus (PVN) of the hypothalamus, which may be responsible for mediating the anorectic effects of NMU (22). It is an intriguing hypothesis that NMUR2 variation may contribute to the BE phenotype in a background of a disrupted neuronetwork and that NMUR2 is possibly one of the candidate or susceptible genes of BE, as shown in rats (10).
Our results also highlight that genetic research will not only assist with the identification of risk alleles but also unlock additional risk factors dependent on genotypes. The advent of next-generation sequencing technologies and its rapid decline in sequencing costs have accelerated the identification of causative variants in most genetic disorders. The rare phenotype in certain diseases may be determined by combined or additive genetic deficits. It is appropriate to define a complete genetic profile of the affected individuals for a phenotype explanation.
In summary, we propose a synergistic genetic effect of NMUR2 and CSF1R variants, which may result in the development of the BE phenotype in the current family with HDLS. Further analysis of NMUR2 in additional BE-related diseases and models would be required to provide supporting evidence.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (NSFC, 81671370; 81471207) and the Program for New Century Excellent Talents in University of China (NCET-12-0069) to WH Xu.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work and for ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. We obtained informed and written consent from available family members, and the study was approved by the Ethical Review Board of Peking Union Medical College Hospital (No. ZS-1554).
References
- Trace SE, Baker JH, Penas-Lledo E, et al. The genetics of eating disorders. Annu Rev Clin Psychol 2013;9:589-620. [Crossref] [PubMed]
- Reichborn-Kjennerud T, Bulik CM, Sullivan PF, et al. Psychiatric and medical symptoms in binge eating in the absence of compensatory behaviors. Obes Res 2004;12:1445-54. [Crossref] [PubMed]
- Johnson JG, Spitzer RL, Williams JB. Health problems, impairment and illnesses associated with bulimia nervosa and binge eating disorder among primary care and obstetric gynaecology patients. Psychol Med 2001;31:1455-66. [Crossref] [PubMed]
- Reichborn-Kjennerud T, Bulik CM, Tambs K, et al. Genetic and environmental influences on binge eating in the absence of compensatory behaviors: a population-based twin study. Int J Eat Disord 2004;36:307-14. [Crossref] [PubMed]
- Javaras KN, Laird NM, Reichborn-Kjennerud T, et al. Familiality and heritability of binge eating disorder: results of a case-control family study and a twin study. Int J Eat Disord 2008;41:174-9. [Crossref] [PubMed]
- Hudson JI, Lalonde JK, Berry JM, et al. Binge-eating disorder as a distinct familial phenotype in obese individuals. Arch Gen Psychiatry 2006;63:313-9. [Crossref] [PubMed]
- Branson R, Potoczna N, Kral JG, et al. Binge eating as a major phenotype of melanocortin 4 receptor gene mutations. N Engl J Med 2003;348:1096-103. [Crossref] [PubMed]
- Cui H, Moore J, Ashimi SS, et al. Eating disorder predisposition is associated with ESRRA and HDAC4 mutations. J Clin Invest 2013;123:4706-13. [Crossref] [PubMed]
- Himmerich H, Bentley J, Kan C, et al. Genetic risk factors for eating disorders: an update and insights into pathophysiology. Ther Adv Psychopharmacol 2019;9:2045125318814734. [Crossref] [PubMed]
- Benzon CR, Johnson SB, McCue DL, et al. Neuromedin U receptor 2 knockdown in the paraventricular nucleus modifies behavioral responses to obesogenic high-fat food and leads to increased body weight. Neuroscience 2014;258:270-9. [Crossref] [PubMed]
- Axelsson R, Roytta M, Sourander P, et al. Hereditary diffuse leucoencephalopathy with spheroids. Acta Psychiatr Scand Suppl 1984;314:1-65. [PubMed]
- Wider C, Van Gerpen JA, DeArmond S, et al. Leukoencephalopathy with spheroids (HDLS) and pigmentary leukodystrophy (POLD): a single entity? Neurology 2009;72:1953-9. [Crossref] [PubMed]
- Wong JC, Chow TW, Hazrati LN. Adult-onset leukoencephalopathy with axonal spheroids and pigmented glia can present as frontotemporal dementia syndrome. Dement Geriatr Cogn Disord 2011;32:150-8. [Crossref] [PubMed]
- Van Gerpen JA, Wider C, Broderick DF, et al. Insights into the dynamics of hereditary diffuse leukoencephalopathy with axonal spheroids. Neurology 2008;71:925-9. [Crossref] [PubMed]
- Rademakers R, Baker M, Nicholson AM, et al. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat Genet 2011;44:200-5. [Crossref] [PubMed]
- Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011;72:257-68. [Crossref] [PubMed]
- Howard AD, Wang R, Pong SS, et al. Identification of receptors for neuromedin U and its role in feeding. Nature 2000;406:70-4. [Crossref] [PubMed]
- Hanada R, Teranishi H, Pearson JT, et al. Neuromedin U has a novel anorexigenic effect independent of the leptin signaling pathway. Nat Med 2004;10:1067-73. [Crossref] [PubMed]
- Konno T, Yoshida K, Mizuta I, et al. Diagnostic criteria for adult-onset leukoencephalopathy with axonal spheroids and pigmented glia due to CSF1R mutation. Eur J Neurol 2018;25:142-7. [Crossref] [PubMed]
- Nicholson AM, Baker MC, Finch NA, et al. CSF1R mutations link POLD and HDLS as a single disease entity. Neurology 2013;80:1033-40. [Crossref] [PubMed]
- Sundal C, Van Gerpen JA, Nicholson AM, et al. MRI characteristics and scoring in HDLS due to CSF1R gene mutations. Neurology 2012;79:566-74. [Crossref] [PubMed]
- Brighton PJ, Szekeres PG, Willars GB. Neuromedin U and its receptors: structure, function, and physiological roles. Pharmacol Rev 2004;56:231-48. [Crossref] [PubMed]


