
Exploration of the relationships between tumor mutation burden with immune infiltrates in clear cell renal cell carcinoma
Introduction
Kidney cancer is the third most common malignance in urinary system, which is only next to prostate cancer and bladder cancer. In 2018, the globally estimated new cases and deaths were 403,262 and 175,098 respectively (1). Renal cell carcinoma (RCC) originates from the renal tubular epithelium, and approximately 90% of kidney tumors belong to RCC. Nowadays, treatments for RCC include surgery with or without postoperative adjuvant therapy, target therapy, chemotherapy and immunotherapy and so on.
The main three histopathological types of RCC includes clear cell RCC (ccRCC), chromophobe RCC (chRCC) and papillary RCC [pRCC, including type I and II (2,3)], and ccRCC accounts for 60–85% in all RCC cases. Due to the lack of reliable diagnostic biomarkers and early clinical symptoms of RCC, over 25% of RCC patients diagnosed with metastatic status, which made it difficult to treat by surgery briefly (4). Generally, ccRCC patients have worse prognosis than chRCC and pRCC (5,6), and the five-year cancer-specific-survival (CSS) for ccRCC for T stage I, II, III and IV was 91%, 74%, 67% and 32% respectively.
Recently, immunotherapy has been recognized to be an effective method for advanced or aggressive cancers (7-9). For example, great efforts have been made in immune checkpoint blockade (ICB) for various malignancies, such as lung cancer (10,11), breast cancer (12,13) and melanoma (14), and the chimeric antigen receptor T cell therapy is being clinical used for leukemia and lymphoma treatment (15). Additionally, many studies had discovered that tumor mutation burden (TMB) and neoepitopes were tightly associated with immunotherapy in many cancer types (16,17). However, only one-fifth of cancer patients could benefit from immunotherapy (18). Therefore, it is of great importance to find the molecular mechanism of immunotherapy responsiveness (19). To date, many molecular determinants have been well identified such as programmed death-ligand-1 (PD-L1) (20), neoantigen load (21), tumor-infiltrating lymphocytes (TILs) (22) and DNA mismatch-repair deficiency (23,24) in many cancers.
With the advanced development of sequencing technique, a lot of valuable bioinformatic sources on TMB and cancer immunotherapeutic response are available from many public databases such as The Cancer Genome Atlas (TCGA) database, Gene Expression Omnibus (GEO) database and so on. Wang’s study showed that the prognostic role of TMB and relationship between TMB and immune infiltrate varied from different types of cancers (25). However, few relevant researches had focused on TMB with immune infiltrates in ccRCC and no accepted opinion had been drawn, so we performed this study to investigate the prognostic role of TMB and the potential association with immune infiltrates in ccRCC.
Methods
Acquisition of somatic mutation data
Somatic mutation data were obtained from the publicly available TCGA database via the GDC data portal (https://portal.gdc.cancer.gov/). From the four subtypes of data files, we selected the “Masked Somatic Mutation” data and processed it based on the VarScan software. We prepared the Mutation Annotation Format (MAF) of somatic variants and implemented the “maftools” (26) R package which provides a multiple of analysis modules to perform the visualization process. Besides, we downloaded the transcriptome profiles with HTSeq-FPKM workflow type of all available ccRCC samples compared with normal tissues. The corresponding clinical information were also obtained from the GDC portal, including clinical variables of age, gender, tumor grade, pathological stage, AJCC-TNM stages and survival outcomes. Since all the data in this research were from public databases, there was no ethical conflict to declare.
Calculation of TMB scores and prognostic analysis
TMB was defined as the total amount of coding errors of somatic genes, base substitutions, insertions or deletions detected across per million bases. In our study, we calculated the mutation frequency with number of variants/the length of exons (38 million) for each sample via Perl scripts based on the JAVA8 platform (http://fp.amegroups.cn/cms/036c91c6905ea44352c2eb045e09f634/atm.2019.10.84-1.pdf). We could classify the ccRCC samples into low- and high-TMB groups according the median data. Then, we merged the TMB data with corresponding survival information via the id number of samples. Kaplan-Meier analysis was conducted to compare the survival difference between low- and high-TMB groups, and the P of log-rank test was calculated. What is more, we further assessed the associations of TMB levels with clinical characteristics, where Wilcoxon rank-sum test was utilized for comparisons between two groups of clinical variables, while Kruskal-Wallis (K-W) test was used when it comes to three or more groups.
Differentially expressed genes and functional pathways analysis
According to the TMB levels, we divided the transcriptome data of ccRCC samples into low- and high-TMB groups via R software. “Limma” was utilized to identify differentially expressed genes (DEGs) in two groups with Fold Change (FC) =2 and False Discovery Rate (FDR) <0.05. Heatmap plot was drawn to show the difference by “pheatmap” package. Then, “org.Hs.eg.db” package was used to get the Entrez ID for each DEG and we conducted the Gene ontology (GO) analysis with “clusterProfiler”, “enrichplot” and “ggplot2” packages. Besides, gene set enrichment analysis (GSEA) was performed based on JAVA8 platform using the TMB level as the phenotype. We selected the “c2.cp.kegg.v6.2.symbols.gmt gene sets” as the reference gene set, which was obtained from the MSigDB database (http://software.broadinstitute.org/gsea/msigdb/). The significant enrichment pathways were considered only with FDR <0.25. Furthermore, we obtained a list of 4677 immune related genes from the Immunology Database and Analysis Portal (Immport) to select the differentially expressed immune genes between the two groups through “VennDiagram” package.
Survival analysis
We selected the top 19 immune genes with |fold change| >2 and FDR <0.05 to further assess the prognostic value of differential immune genes in patients with low- and high-TMB levels. Bath survival Kaplan-Meier analysis was conducted via a “for cycle” R script to find the hub immune genes associated with survival outcomes and the P value was shown in plot. A P value <0.05 was significant.
TIMER database and CIBERSORT algorithm
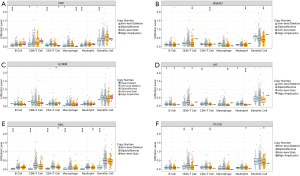
We further evaluated the mutation types of hub immune genes with immune infiltrates in ccRCC based on the “SCNA” module of TIMER database (27) (https://cistrome.shinyapps.io/timer/). The known mutation types of 6 hub genes were shown at the right bottom. The distributions of each immune cell subset at each mutation status in ccRCC were presented by box plots and the difference of infiltration level in each category versus normal was compared using two-sided Wilcoxon rank sum test with calculated P value.
Meanwhile, we obtained the transcriptome profiles of ccRCC patients in two groups and conducted the normalization process via “limma” package. Then, we put the preparation data into subsequent analysis to assess the immune infractions of each sample through the CIBERSORT algorithm (R script v1.03), providing an estimation of the abundances of member cell types in a mixed cell population, using gene expression data. The CIBERSORT was still based on a known reference set, providing a set of gene expression features of 22 leukocyte subtypes-LM22. The distributions of immune cells in two groups were shown by “pheatmap” package. Wilcoxon rank-sum test was exploited to compare the differential abundances of immune infiltrates between low- and high-TMB groups, which were exhibited with P value by “vioplot” package.
Prognostic analysis of immune cells in ccRCC
Based on the TIMER database, we further conducted the multivariate Cox of immune infiltration cells, which was fitted by function coxph() from R package “survival”. The hazard ratio (HR) with 95% confidence interval (95%CI) was calculated. Furthermore, the Kaplan-Meier analysis was conducted to show the differential survival outcomes between different levels of immune infiltrates. A P value <0.05 of log-rank test was regarded as the statistical significance.
Construction of TMB Prognostic index (TMBPI) for hub immune genes
We conducted the multivariate Cox regression analysis to obtain the respective coefficients (βi) of 9 hub immune genes. The TMBPI was constructed as: TMBPI = Ʃ(βi × Expi) (i=9). Then, we performed the Receiver Operating Characteristic (ROC) curve to assess the predictive value of 9 immune signature in ccRCC. Moreover, Kaplan-Meier analysis was conducted to compare the survival difference in two groups, where we divided the 530 ccRCC patients into high- and low-groups with the median prognostic index as the threshold.
Statistical analysis
The Cox regression model was performed based on the “survival” package. “Limma” package was utilized to conduct the normalization and differential analysis. Wilcoxon rank-sum test was a non-parametric statistical hypothesis test mainly used for comparisons between two groups and Kruskal-Wallis test was suitable for two or more categories. All statistical analysis was implemented based on the R software (Version 3.5.2). A P value <0.05 was thought to be significant.
Results
Landscape of mutation profiles in ccRCC
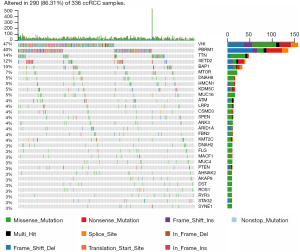
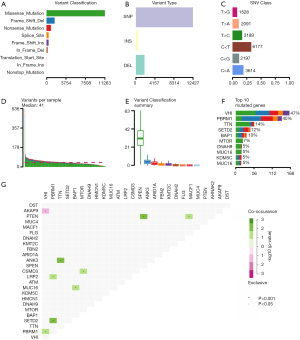
We downloaded the somatic mutation profiles of 339 ccRCC patients from TCGA, including four types of data based on diverse processing software. We utilized the “maftools” package to visualize the results based on the mutation data with VCF format. Mutation information of each gene in each sample was exhibited in waterfall plot, where various colors with annotations at the bottom represented the different mutation types (Figure 1). In summary, these mutations were further classified according to different classified categories, in which missense mutation accounts for the most fraction (Figure 2A), single nucleotide polymorphism occurred more frequently than insertion or deletion (Figure 2B), and C>T was the most common of single nucleotide variants (SNV) in ccRCC (Figure 2C). Besides, we counted the number of altered bases in each sample and showed the mutation type with different colors in box plot for ccRCC (Figure 2D,E). Last, we exhibited the top 10 mutated genes in ccRCC with ranked percentages, including VHL (47%), PBRM1 (40%), TTN (14%), SETD2 (12%), BAP1 (10%), MTOR (7%), DNAH9 (5%), MUC16 (5%), KDM5C (5%) and HMCN1 (5%) (Figure 2F). The coincident and exclusive associations across mutated genes were shown in Figure 2G, where green represented the co-occurrence and red represented the mutually exclusive relationships. Meanwhile, the mutated frequencies of other genes were shown by Genecloud plot in Figure S1.
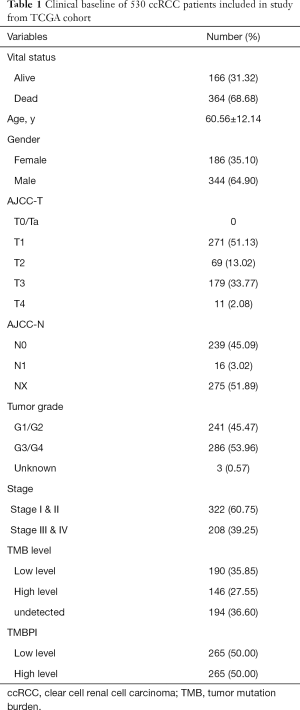
Moreover, the transcriptome profiles were obtained consisted of 539 ccRCC and 72 normal samples. The clinical information of 530 ccRCC patients were shown in Table 1, in which the mean age of patients was 60.56±12.14 years old and they contained 344 males and 186 females.
Full table
TMB correlated with survival outcomes, pathological stages and tumor grades
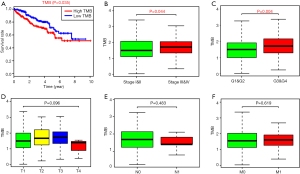
We calculated the amount of mutation events per million bases as the TMB for 336 ccRCC, and we further divided the patients into two groups with high- and low-TMB levels using the median TMB as the cutoff value (http://fp.amegroups.cn/cms/036c91c6905ea44352c2eb045e09f634/atm.2019.10.84-1.pdf). Contrary to other conditions that higher TMB tends to promote immune recognition and results in better prognosis, patients in high-TMB group revealed worse survival outcomes than that in low-TMB group with log-rank test of P=0.035 (Figure 3A). Furthermore, higher TMB levels correlated with advanced pathological stages with P=0.044 (Figure 3B), and higher tumor grades with P=0.004 (Figure 3C). However, no significant differences were observed in associations of TMB with AJCC-T, N, M stages (Figure 3D,E,F).
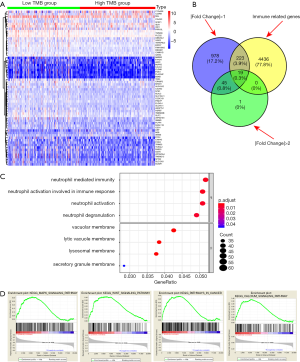
Comparison of gene expression profiles between low- and high-TMB groups
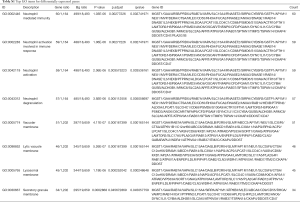
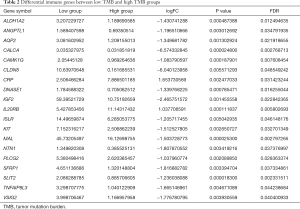
The heatmap indicated that genes in high-TMB group showed commonly lower expression levels than that in low-TMB group (Figure 4A). Differential analysis revealed a list of 1,265 DEGs with |Fold Change| >1 and 65 DEGs with |Fold Change| >2 in Venn plot (Figure 4B, http://fp.amegroups.cn/cms/89d685ac9955df9b8c0a5ef77f653065/atm.2019.10.84-2.pdf). We then conducted the GO enrichment analysis and these DEGs were mainly involved in neutrophil mediated immunity, neutrophil activation and other immune-related crosstalk. In addition, we further selected the GSEA results of the top TMB-related items in Figure 4D, including MAPK signaling pathway, Wnt signaling pathway, pathway in cancers and calcium associated crosstalk with FDR <0.25 (Figure 4D, Table S1). Since the TMB correlated with immune signature or pathways in ccRCC, we further identified top 19 immune related genes from Immport database for subsequent analysis (Table 2).
Full table
Full table
Identification of hub TMB-related immune genes and associations of mutants with immune infiltrates
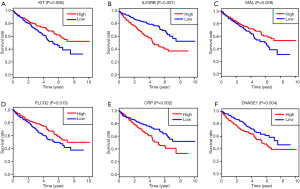
Batch survival analysis was conducted with R scripts to screen 6 prognostic hub immune genes that highly associated with survival. Higher expression levels of IL20B, CRP and DNASE1 correlated positively with poor survival outcomes, while expression level of KIT, MAL and PLCG2 correlated negatively with prognosis (Figure 5). More importantly, we further assessed the underlying relationships of the mutants of these hub genes with immune infiltrates in ccRCC microenvironment. Compared with the immune infiltration levels in samples with wild type of the signature, diverse forms of mutation carried by hub genes could commonly inhibit the immune infiltrates, including CD8+ T cell, neutrophil cell, dendritic cell, macrophage, CD4+ T cell and B cell (Figure 6).
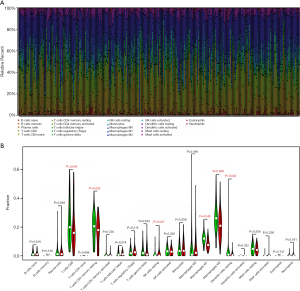
Differential abundance of immune cells in low- and high-TMB groups
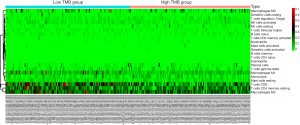
Since we have demonstrated that the DGEs were involved in immune crosstalk and mutants of hub immune signature negatively correlated with immune infiltrates, we intended to further compare the differential profiles of immune fractions between high- and low-TMB groups (http://fp.amegroups.cn/cms/4f8c1ec9c0638cb1728c073c3a3dc64a/atm.2019.10.84-3.pdf). After the filtration of samples with P>0.05 via “CIBERSORT” package, a total of 238 samples were selected to perform the immune cells analysis, including 121 low-TMB samples and 117 high-TMB samples. Specific fractions of 22 immune cells in each ccRCC sample were shown by box plot in Figure 7A. Besides, Wilcoxon rank-sum test revealed that the infiltration levels of CD8+ T cell, CD4+ memory resting T cell, M1 and M2 macrophages, as well as dendritic resting cells in high-TMB group were lower compared with that in low-TMB group (Figure 7B). What is more, we revealed the distributions of 22 immune cells fractions in two groups by heatmap plot (Figure S2). In accordance with the previous mutation analysis, higher TMB commonly inhibited the immune infiltration levels in ccRCC samples.
Low CD8+ T cell and macrophage infiltrates confers poor survival outcomes
To further investigate the underlying prognosis of immune cells, we further conducted the Cox regression model in ccRCC samples and the model was constructed as: Surv(KIRC) ~ B cell + CD8+ T cell + Macrophage + CD4+ T cell + Dendritic cell + Neutrophil cell. The Cox results suggested that lower CD8+ T cell (HR =0.175, P=0.029) and Macrophage (HR =0.062, P=0.020) infiltrates were risk factors for ccRCC (Table 3). Furthermore, we performed the Kaplan-Meier analysis, in which lower infiltration levels of CD8+ T cell and Macrophage indeed correlated with poor survival outcomes in ccRCC (Figure 8A).

Full table
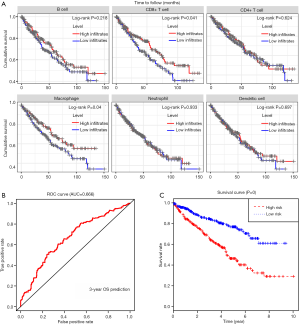
Construction and assessment of TMBPI for ccRCC
Based on the above analysis that the alteration of immune signature led to lower immune infiltrates and poor prognosis, we wanted to evaluate the predictive accuracy of the 6 TMB-related hub signature. We extracted the transcriptome data from 530 ccRCC matched with compete clinical information via “merge” function in R studio. Based on the multivariate Cox regression model, we constructed the TMBPI as the following: PI = (0.042086 × CRP + 0.347889 × DNASE1 + 0.118190 × IL20RB − 0.027266 × KIT − 0.007359 × MAL − 0.256051 × PLCG2). Then, we calculated the TMBPI for each ccRCC patients and divided them into high- (n=265) and low-risk level (n=265) with the median cutoff value (http://fp.amegroups.cn/cms/89d685ac9955df9b8c0a5ef77f653065/atm.2019.10.84-2.pdf). The ROC curve of 3-year OS prediction was drawn to assess the predictive accuracy with AUC =0.666 (Figure 8B). Meanwhile, Kaplan-Meier plot showed that patients with high TMBPI revealed worse survival outcomes compared with that with low TMBPI, which deserved further large samples to validate (Figure 8C).
Discussion
ccRCC is the most common pathological type of RCC. In ccRCC, the von Hippel-Lindau (VHL) gene mutation and chromosome 3p loss are always founded, including some additional tumor suppressor genes which are frequently identified next to the VHL gene, such as PBRM 1, SETD 2 and BAP1 et al. (28). Immunotherapy in the treatment of advanced or aggressive cancers has shown promising results (29,30). Nowadays, immunotherapy for RCC is changing from conventional IL2 non-specific immunotherapy to specific targeted immunotherapy based on programmed death-1 (PD-1) and PD-L1 inhibitors at immune checkpoints. However, only a fraction of patients benefit from immunotherapy while most patients have no immune response, which represents terrible waste of money. Hence, identifying patients who are more likely to respond to immunotherapy before taking the drugs seems to be particularly important.
Recently, the application of high-throughput sequencing technology has brought up several genome-wide biomarkers for ccRCC, such as chromosomal abnormalities which was reported to be independently associated with worse outcomes. Accordingly, TMB detected from genome-sequencing is a novel biomarker to predict response to ICB, which has been demonstrated its effectiveness in a variety of tumors, such as breast cancer (31), lung cancer (32) and so on. Alexandra Thomas (33) demonstrated that TMB was a determinant of immune-related survival in breast cancer patients and can be a useful tool to identify candidate immunoregulatory mechanisms associated with immunological cold tumors. Negrao (34) found that low TMB, negative PD-L1 expression, presence of targetable driver mutation and serine/threonine kinase 11 gene (STK11) mutation were predictive factors of worse outcome in non-small cell lung cancer. However, its prognostic role and association with immunotherapy in ccRCC have not been explored, so we conduct this research investigate the prognostic role of TMB and the potential association with immune infiltrates in ccRCC.
Based on the results of the survival analysis, patients in High-TMB group showed worse survival outcomes, advanced pathological stages and higher tumor grades than that in low-TMB group. Pai et al. (35) discovered that there was a trend towards improved progression free survival (PFS) in Low-TMB colorectal cancer patients (n=39) compared to High-/Intermediate-TMB (n=26) (9.9 vs. 5.8 months), but did not reach statistical significance (P=0.18). However, Goodman et al. (36) found that melanoma and non-small cell lung cancer patients with High-TMB revealed better PFS than that with Low-TMB. Similar conclusion was drawn by Park (31) that high TMB in patients with HER2-positive refractory metastatic breast cancer may be a prognostic marker for good overall survival.
DEGs were ferreted out and GO enrichment analysis showed that these DEGs were mainly involved in neutrophil mediated immunity, neutrophil activation and other immune-related crosstalk. Hence, 6 prognostic hub immune genes that highly associated with survival were identified (positive correlation: IL20B, CRP and DNASE1, negative correlation: KIT, MAL and PLCG2). Moreover, mutants of these hub immune genes were related to immune infiltrates in ccRCC microenvironment. Immune infiltrates including CD8+ T cell, neutrophil cell, dendritic cell, macrophage, CD4+ T cell and B cell could be inhibited by hub genes mutations.
KIT is a proto-oncogene which encodes a member of the type III receptor tyrosine kinase family (37,38) and KIT activating mutations lead to dysregulation of downstream signaling pathways of hematopoietic stem cells for the survival, proliferation and differentiation. Previous studies had demonstrated that KIT mutations may result in adverse survival in acute myeloid leukemia (39,40) and gastroenteropancreatic neuroendocrine tumor (41), however, in vulvar melanomas KIT mutations are markers of better PFS (42). MAL encodes a membrane protein involved in lipid raft organization during T-cell signal transduction and activation (43,44), and MAL overexpression was associated with poor survival in ovarian carcinoma (45) and Hodgkin lymphoma (46).
In the analysis of underlying prognosis of immune cells, lower CD8+ T cell and macrophage infiltrates were proven to be tightly associated with poor survival outcomes. Yao et al. (47) found that metastatic RCC patients with high CD8+ T lymphocytes infiltration and low PD-1 expression had longer survival. Also, similar result can be found that high grade serous ovarian cancer patients with lower numbers of intraepithelial CD8+ TILs and positive tumor PD-L1 expression had the shortest median OS (48). Moreover, previous relevant studies (49,50) had also explored the prognostic role of macrophage infiltrates in immunotherapy, tumor infiltrating macrophages could play an important role in metastasis suppression progress in metastatic diseases.
Finally, a prognostic model (TMBPI) was developed using 6 hub immune genes which can be great useful for survival prediction. To our knowledge, this is the first TMB prognostic model to predict survival outcomes. Patients with high TMBPI revealed worse survival outcomes compared with that with low TMBPI. However, the AUC of this predictive model was only 0.666 and further large-sample researches are needed for verification and modification before clinical application.
However, there are some limitations that should not been ignored: (I) lack of basic experiment to validate the association between 6 immune genes signature and tumor cell immune infiltrates; (II) lack of large clinical sample to verify the prognostic effect of TMB and its potential relationship with immune infiltrates. Relevant variants and big sample clinical trials are needed in the future.
Conclusions
Higher TMB correlated with poor survival outcomes and might inhibit the immune infiltrates in ccRCC. The mutants of 6 hub TMB-related immune signature conferred lower immune cells infiltration which deserved further validation.
Acknowledgments
Funding: The Training Program of Outstanding Young Talents in Clinical Medicine by Hongkou District, Grant/Award Number: No. HKYQ2018-09.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Delahunt B, Eble JN. Papillary renal cell carcinoma: a clinicopathologic and immunohistochemical study of 105 tumors. Mod Pathol 1997;10:537-44. [PubMed]
- Delahunt B, Eble JN, McCredie MR, et al. Morphologic typing of papillary renal cell carcinoma: comparison of growth kinetics and patient survival in 66 cases. Hum Pathol 2001;32:590-5. [Crossref] [PubMed]
- Meskawi M, Sun M, Trinh QD, et al. A review of integrated staging systems for renal cell carcinoma. Eur Urol 2012;62:303-14. [Crossref] [PubMed]
- Capitanio U, Cloutier V, Zini L, et al. A critical assessment of the prognostic value of clear cell, papillary and chromophobe histological subtypes in renal cell carcinoma: a population-based study. BJU Int 2009;103:1496-500. [Crossref] [PubMed]
- Keegan KA, Schupp CW, Chamie K, et al. Histopathology of surgically treated renal cell carcinoma: survival differences by subtype and stage. J Urol 2012;188:391-7. [Crossref] [PubMed]
- Kamal Y, Cheng C, Frost HR, et al. Predictors of disease aggressiveness influence outcome from immunotherapy treatment in renal clear cell carcinoma. Oncoimmunology 2018;8:e1500106. [Crossref] [PubMed]
- Hoos A. Development of immuno-oncology drugs - from CTLA4 to PD1 to the next generations. Nat Rev Drug Discov 2016;15:235-47. [Crossref] [PubMed]
- Aoun F, Rassy EE, Assi T, et al. Advances in urothelial bladder cancer immunotherapy, dawn of a new age of treatment. Immunotherapy 2017;9:451-60. [Crossref] [PubMed]
- Yang J, Chen J, Wei J, et al. Immune checkpoint blockade as a potential therapeutic target in non-small cell lung cancer. Expert Opin Biol Ther 2016;16:1209-23. [Crossref] [PubMed]
- Anagnostou V, Smith KN, Forde PM, et al. Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Discov 2017;7:264-76. [Crossref] [PubMed]
- Bu X, Yao Y, Li X. Immune Checkpoint Blockade in Breast Cancer Therapy. Adv Exp Med Biol 2017;1026:383-402. [Crossref] [PubMed]
- Hu ZI, Ho AY, McArthur HL. Combined Radiation Therapy and Immune Checkpoint Blockade Therapy for Breast Cancer. Int J Radiat Oncol Biol Phys 2017;99:153-64. [Crossref] [PubMed]
- Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207-11. [Crossref] [PubMed]
- June CH, O'Connor RS, Kawalekar OU, et al. CAR T cell immunotherapy for human cancer. Science 2018;359:1361-5. [Crossref] [PubMed]
- Brown SD, Warren RL, Gibb EA, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res 2014;24:743-50. [Crossref] [PubMed]
- Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333-9. [Crossref] [PubMed]
- Braun DA, Burke KP, Van Allen EM. Genomic Approaches to Understanding Response and Resistance to Immunotherapy. Clin Cancer Res 2016;22:5642-50. [Crossref] [PubMed]
- Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol 2016;17:e542-51. [Crossref] [PubMed]
- Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther 2015;14:847-56. [Crossref] [PubMed]
- Efremova M, Finotello F, Rieder D, et al. Neoantigens Generated by Individual Mutations and Their Role in Cancer Immunity and Immunotherapy. Front Immunol 2017;8:1679. [Crossref] [PubMed]
- Zito Marino F, Ascierto PA, Rossi G, et al. Are tumor-infiltrating lymphocytes protagonists or background actors in patient selection for cancer immunotherapy? Expert Opin Biol Ther 2017;17:735-46. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Diaz LA Jr, Le DT. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;373:1979. [Crossref] [PubMed]
- Wang X, Li M. Correlate tumor mutation burden with immune signatures in human cancers. BMC Immunol 2019;20:4. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Brugarolas J. Molecular genetics of clear-cell renal cell carcinoma. J Clin Oncol 2014;32:1968-76. [Crossref] [PubMed]
- Voena C, Chiarle R. Advances in cancer immunology and cancer immunotherapy. Discov Med 2016;21:125-33. [PubMed]
- Menon S, Shin S, Dy G. Advances in Cancer Immunotherapy in Solid Tumors. Cancers (Basel) 2016.8. [PubMed]
- Park SE, Park K, Lee E, et al. Clinical implication of tumor mutational burden in patients with HER2-positive refractory metastatic breast cancer. Oncoimmunology 2018;7:e1466768. [Crossref] [PubMed]
- Hellmann MD, Callahan MK, Awad MM, et al. Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in Combination with Ipilimumab in Small-Cell Lung Cancer. Cancer Cell 2018;33:853-61.e4. [Crossref] [PubMed]
- Thomas A, Routh ED, Pullikuth A, et al. Tumor mutational burden is a determinant of immune-mediated survival in breast cancer. Oncoimmunology 2018;7:e1490854. [Crossref] [PubMed]
- Negrao MV, Lam VK, Reuben A, et al. PD-L1 Expression, Tumor Mutational Burden, and Cancer Gene Mutations Are Stronger Predictors of Benefit from Immune Checkpoint Blockade than HLA Class I Genotype in Non-Small Cell Lung Cancer. J Thorac Oncol 2019;14:1021-31. [Crossref] [PubMed]
- Pai SG, Carneiro BA, Chae YK, et al. Correlation of tumor mutational burden and treatment outcomes in patients with colorectal cancer. J Gastrointest Oncol 2017;8:858-66. [Crossref] [PubMed]
- Goodman AM, Kato S, Bazhenova L, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther 2017;16:2598-608. [Crossref] [PubMed]
- d'Auriol L, Mattei MG, Andre C, et al. Localization of the human c-kit protooncogene on the q11-q12 region of chromosome 4. Hum Genet 1988;78:374-6. [Crossref] [PubMed]
- Paschka P, Marcucci G, Ruppert AS, et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B Study. J Clin Oncol 2006;24:3904-11. [Crossref] [PubMed]
- Chen X, Dou H, Wang X, et al. KIT mutations correlate with adverse survival in children with core-binding factor acute myeloid leukemia. Leuk Lymphoma 2018;59:829-36. [Crossref] [PubMed]
- Allen C, Hills RK, Lamb K, et al. The importance of relative mutant level for evaluating impact on outcome of KIT, FLT3 and CBL mutations in core-binding factor acute myeloid leukemia. Leukemia 2013;27:1891-901. [Crossref] [PubMed]
- Knösel T, Chen Y, Altendorf-Hofmann A, et al. High KIT and PDGFRA are associated with shorter patients survival in gastroenteropancreatic neuroendocrine tumors, but mutations are a rare event. J Cancer Res Clin Oncol 2012;138:397-403. [Crossref] [PubMed]
- Dias-Santagata D, Selim MA, Su Y, et al. KIT mutations and CD117 overexpression are markers of better progression-free survival in vulvar melanomas. Br J Dermatol 2017;177:1376-84. [Crossref] [PubMed]
- Alonso MA, Millan J. The role of lipid rafts in signalling and membrane trafficking in T lymphocytes. J Cell Sci 2001;114:3957-65. [PubMed]
- Copie-Bergman C, Plonquet A, Alonso MA, et al. MAL expression in lymphoid cells: further evidence for MAL as a distinct molecular marker of primary mediastinal large B-cell lymphomas. Mod Pathol 2002;15:1172-80. [Crossref] [PubMed]
- Berchuck A, Iversen ES, Lancaster JM, et al. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin Cancer Res 2005;11:3686-96. [Crossref] [PubMed]
- Hsi ED, Sup SJ, Alemany C, et al. MAL is expressed in a subset of Hodgkin lymphoma and identifies a population of patients with poor prognosis. Am J Clin Pathol 2006;125:776-82. [Crossref] [PubMed]
- Yao J, Xi W, Zhu Y, et al. Checkpoint molecule PD-1-assisted CD8(+) T lymphocyte count in tumor microenvironment predicts overall survival of patients with metastatic renal cell carcinoma treated with tyrosine kinase inhibitors. Cancer Manag Res 2018;10:3419-31. [Crossref] [PubMed]
- Wang Q, Lou W, Di W, et al. Prognostic value of tumor PD-L1 expression combined with CD8(+) tumor infiltrating lymphocytes in high grade serous ovarian cancer. Int Immunopharmacol 2017;52:7-14. [Crossref] [PubMed]
- Withers SS, Skorupski KA, York D, et al. Association of macrophage and lymphocyte infiltration with outcome in canine osteosarcoma. Vet Comp Oncol 2019;17:49-60. [Crossref] [PubMed]
- Buddingh EP, Kuijjer ML, Duim RA, et al. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: a rationale for treatment with macrophage activating agents. Clin Cancer Res 2011;17:2110-9. [Crossref] [PubMed]


