
CDK14 involvement in proliferation migration and invasion of esophageal cancer
Introduction
Esophageal cancer is the eighth most common cancer in the world (1). It has a high lethality rate, with more than 400,000 deaths per year (2,3). Esophageal squamous cell carcinoma (ESCC) is the most common type of esophageal carcinoma globally and in economically developing countries. Approximately half of the world’s 500,000 new ESCC cases annually occur in China, where the disease is a major public health problem (4). With insidious early symptoms, patients with ESCC often have advanced stage disease at initial diagnosis. As an effective means of comprehensive therapy, chemotherapy is important in esophageal cancer treatment. Responses to chemotherapy differ widely and have considerable heterogeneity in patients. The resistance of tumor cells to chemotherapy drugs restricts treatment effects and results in poor prognosis. Therefore, research on molecular mechanisms related to chemoresistance is essential to reduce the occurrence and development of esophageal cancer.
Cyclin-dependent kinases (CDKs) are a family of 20 protein kinases that regulate cell cycle progression (Cdk1, Cdk2, Cdk3, Cdk4, and Cdk6), transcription (Cdk7, Cdk8, Cdk9, and Cdk10), differentiation (Cdk5), and other processes (5-7). CDK14, also known as PFTAIRE1, is a novel member of the Cdc2-related serine/threonine protein kinases and the CDK family (8,9). Studies show that CDK14 regulates cell cycle, cell proliferation and several pathways and is an oncogene in a variety of human malignant cancers (10,11). For instance, CDK14 interacts with cyclin Y to activate noncanonical Wnt signaling in hepatocellular carcinoma (12). Knockdown of CDK14 inhibits tumor cell proliferation, invasion and epithelial-to-mesenchymal transition in pancreatic cancer. The abnormal expression of CDK14 is related to chemoresistance in patients with oesophageal squamous cell carcinoma and could be useful as a prognostic marker (13). How CDK14 is involved in ESCC is not clear.
Our work explored the concrete mechanism of CDK14 in proliferation of ESCC. Western blots and immunohistochemistry (IHC) analysis verified that expression of CDK14 in tumor tissues is increased which is compared to it in normal tissues and cell lines IHC staining revealed that CDK14 positively correlated with clinical pathological variables such as tumor size, tumor grade and tumor invasion, and Ki-67. Protein spectrum analysis suggests that CDK14 interacts with CDK7/CCNH. In vitro, starvation and refeeding assays demonstrated that CDK14 expression was increased and was co-located with CDK7 in ESCC cell. It has been proved that CDK7/CCNH is a CAK complex that is implicated in activating several types of CDKs including CDK2, CDK4 by phosphorylation. We found that the CAK complex (CDK7/CCNH)/CDK14 had physical interactions in ECA109 cells. Through overexpression experiments for pathways, we showed that CDK14 was activated by the CAK complex and increased proliferation in ESCC, which reduced the sensitivity of ESCC cell to cisplatin.
Methods
Patients and tissue samples
Eight pairs of fresh tissues from ESCC tumors and matched adjacent normal esophageal epithelia were stored at −80 °C immediately after surgical removal and before western blot analysis. One hundred and three ESCC specimens was provided by the Department of Pathology, Affiliated Hospital of Nantong University to conduct IHC analysis from 2008 to 2014. Patients were excluded if they had immunotherapy, radiation, or chemotherapy as preoperative therapies. All patients were followed for 1 to 60 months. For histological examinations, all tumorous and surrounding nontumorous tissues were fixed in 10% buffered formalin and embedded in paraffin for sectioning. Resected specimens were classified using the TNM classification system. The Ethics Committee of the Affiliated Hospital of Nantong University permitted use of the tissue sections for research purposes. Signed informed consent was obtained from the 103 patients. Main clinical and pathological variables are in Table 1.
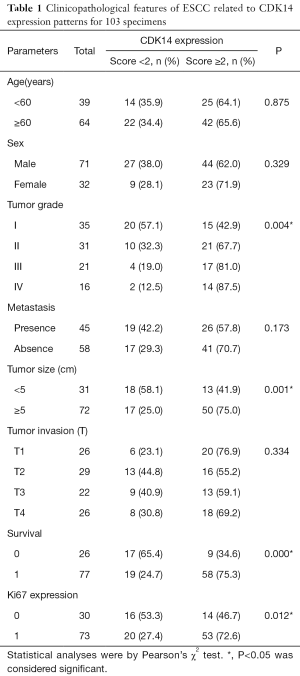
Full table
Western blots
Tissues and harvested cells were homogenized in homogenization buffer containing 1 M Tris-HCl, pH 7.5, 1% TritonX-100, 1% Nonidet P-40, 10% sodium dodecyl sulfate (SDS), 0.5% sodium deoxycholate, 0.5 M EDTA, 10 µg/mL leupeptin, 10 µg/mL aprotinin, and 1 mM PMSF, then centrifuged at 13,000× g for 15 min to collect supernatant. Protein concentration was determined with a Bio-Rad protein assay (Bio-Rad, Hercules, CA, USA). Supernatant was diluted in 2× SDS loading buffer and boiled. Equal amounts of protein were separated by 10% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and transferred to PVDF membranes (Millipore, Bedford, MA, USA). After blocking with 5% nonfat milk or bovine serum albumin (BSA) or 2 h at room temperature, membranes were incubated overnight at 4 °C with primary antibodies: (I) anti-CDK14 (1:500, BD Biosciences), (II) anti-CDK7 (1:1,000, Santa Cruz Biotechnology), (III) anti-CCNH (1:1,000, Santa Cruz Biotechnology), (IV) anti-GAPDH (1:3,000, Sigma-Aldrich), (V) anti-PCNA (1:1,000, Santa Cruz Biotechnology), (VI) anti-cyclin E (1:500, Santa Cruz Biotechnology), (VII) anti-Thr (1:500, Santa Cruz Biotechnology), (VIII) anti-Ser (1:500, Santa Cruz Biotechnology), (IX) anti-Rb (Cell Signaling Technology). After three washes, membranes were incubated with horseradish peroxidase-linked secondary human anti-mouse or anti-rabbit antibodies (1:5,000; Pierce Biotechnology) for 2 h at room temperature. Detection of immunocomplexes was by scanning using a Molecular Dynamics densitometer (Imaging Technology, Ontario, Canada). Image J (National Institutes of Health) was used to measure band density. Experiments were carried out on three separate occasions.
Immunohistochemical staining
Serial sections of 5 µm were mounted on glass slides coated with 10% polylysine, dewaxed in xylene, and rehydrated through graded alcohol. Sections were processed in 10 mM citrate buffer (Ph 6.0) and heated to 121 °C in an autoclave for 3 min to retrieve antigen. Endogenous peroxidase activity was blocked by 3% hydrogen peroxide for 15 min. Antibodies used were anti-CDK14 (1:50, BD Biosciences) and anti-ki-67 (1:400, Santa Cruz Biotechnology). Negative control slides were treated in parallel using nonspecific immunoglobulin IgG (Sigma-Aldrich) at the same concentration as the primary antibody. Finally, DAB was used to visualize immunoreactivity and counterstained with hematoxylin. All slides were processed using the avidin-biotin-peroxidase complex method. Antigen-antibody reactions were visualized with chromogen diaminobenzidine.
Immunohistochemical evaluation
Three pathologists blinded to histopathological features and patient data for samples independently evaluated the degree of immunostaining. For statistical analysis of CDK14 staining, five high-power fields were randomly chosen, and at least 300 cells were counted per field. Expression scores were determined by staining intensity and immunoreactive cell percentage. Tissues with no staining were rated 0, faint staining or moderate to strong staining in <25% of cells were 1, moderate staining or strong staining in 25–50% of cells were 2, and strong staining in ≥50% of cells was 3. For statistical analysis, <2 was low expression and ≥2 was overexpression. For all sections, cell counting was repeated three times to avoid technical errors; similar results were obtained for samples.
Cell culture and transient transfection
IHC stain was as previously described (14). Human esophageal cancer cell line Eca109 was from the China Academy of Science cell library, and was cultured at 37 °C and 5% CO2 in RPMI-1640 medium (Gibco BRL, Grand Island, NY, USA) with 10% heat-inactivated fetal bovine serum (Hyclone, Logan, UT, USA) supplemented with 100 U/mL penicillin–streptomycin mixture (Gibco BRL, Grand Island, NY, USA) and 2 mM L-glutamine. Before transfection, medium was renewed after 24 h by fresh one. Transfection used Super Fectin transfection reagent (Invitrogen) according to the manufacturer’s instructions. Transfected cells were harvested 48 h after transfection and used for western blots, co-immunoprecipitation, colony formation, and flow cytometric assays.
Co-immunoprecipitation and immunofluorescence assays
Eca109 cells were washed three times with cold phosphate-buffered saline (PBS). Cells were lysed in plates with 1 mL radioimmunoprecipitation assay buffer (150 MM NaCl, 0.5% Nonidet P-40, 50 m
Flow cytometric analysis
Flow cytometric analysis was as previously described (15). For cell-cycle detection, a single-cell suspension was fixed with 70% ethanol at −20 °C for 24 h. Cells were incubated with 1 mg/mL RNase A in PBS at 37 °C for 30 min and stained with 0.5 mg/mL propidium iodide. DNA contents were analyzed by a flow cytometer (Becton Dickinson, Lincoln Park, NJ, USA). To detect apoptosis, cells were seeded onto 6-well plates with 104 cells/well in 1.5 mL culture medium and allowed to adhere overnight before transfection. At 24 to 36 h after transfection, medium was replaced with LY294002. After 48 h incubation, cells were harvested and apoptosis was determined by flow cytometry using Annexin V-PE/7-AAD and Annexin V-FITC/PI apoptosis detection kits (KeyGen Biotechnology Co., Ltd, Nanjing, China) according to the manufacturer’s instructions. Three independent experiments were conducted.
Colony formation assays
ECA109 cells were plated in 6-well plates (0.5×103 cells each plate) following specific transfection. After 10–14 days, surviving colonies were stained with 0.5% Crystal Niolet and ≥50 cells/colony were counted. Three independent experiments were conducted.
Proliferation assays
Cell proliferation was measured using commercial Cell Counting Kits (CCK)-8 (Dojindo, Kumamoto, Japan) following the manufacturer’s instructions. ECA109 cells were seeded onto 96-well culture cluster plates (Corning, NY, USA) at 2×104 cells/well in 1 mL culture medium, and grown overnight. Kit reagents were added to a subset of wells for 2 h at 37 °C and absorbance was read with an automated plate reader after treatment. Three independent experiments were conducted.
Mammalian expression constructs
Human CDK7 and CCNH cDNA was from Genechem (Shanghai, China). Human CDK14 cDNA was from Generay Biotechnology (Shanghai, China). Derivative cDNAs were constructed into a N-Terminal p3xFLAG-CMV vector which was purchased from Sigma-Aldrich (Shanghai, China). PCR primers were: CDK14-F, 5'-CGGAATTCAATGTGTGACCTCATTGAG-3' and CDK14-R, 3'-GGGGTACCAGTGCTTGCTGTTTGATAG-5', N1-F, 5'-CGGAATTCAATGGTTAGGCGGCAC-3' and N1-R, 3'-GGGGTACCAGTGCTTGCTGTTTGATAG-5', C1-F, 5'-CGGAATTCAATGTGTGACCTCATTGAG-3', and C1-R, 3'-GGGGTACCATGAGTCAGCTTTTCC-5'. PCR was by SYBR Premix Ex Taq Kits and an Applied Biosystems 7500HT Fast RealTime PCR System. PCR product specificity was checked by melting temperatures and curves.
Statistical analysis
All computations were performed by a State 8.0 software package. Results were expressed as mean ± SEM. Associations between CDK14 expression and clinicopathological features were analyzed using χ2 tests. For analysis of survival data, Kaplan-Meier curves were constructed, and log-rank tests were performed. Multivariate analysis was by a Cox’s proportional hazards model, and risk ratios and 95% confidence intervals were recorded for all markers. All data represent the results of at least three independent experiments at least and P values <0.05 were considered significant.
Results
CDK14 was highly expressed in ESCC tissues and cells
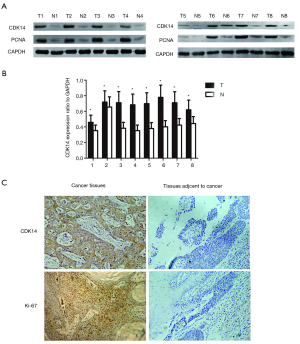
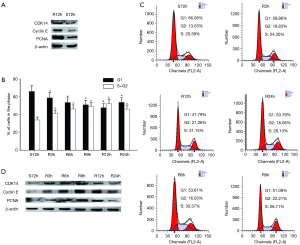
In previous work, we found elevated CDK7 and CCNH protein levels in ESCC tissues compared with adjacent normal tissues. Low expression decreased proliferation of human ESCC (16). To assess expression of CDK14 and proliferating cell nuclear antigen (PCNA) in ESCC again, we performed western blots using eight paired ESCC tumors and matched adjacent normal esophageal epithelia fresh frozen tissues and the Eca109 cell line. PCNA is a marker of increased cell proliferation. CDK14 expression was significantly higher in ESCC tissues than normal tissues via statistical comparison of the grey value (Figure 1A,B). The expression pattern of CDK14 paralleled PCNA. IHC analyses were used to detect CDK14 expression in 103 ESCC samples. CDK14 was diffusely located in the cytoplasm and nucleus, while Ki-67 (a cell proliferation index) was mainly in the nucleus (Figure 1C).
CDK14 expression, pathological parameters and prognostic significance
To determine the clinical significance in ESCC, we used Pearson’s χ2 test to assess association of CDK14 expression with clinical-pathological parameters and ki-67. Using CDK14 immunostaining scores, patient samples were divided into positive and negative groups. Expression of CDK14 was related to tumor grade (P=0.001), tumor size (P=0.001), tumor invasion (P=0.025), survival (P=0.000) and Ki-67 (P=0.012) (Table 1). No relationship was seen between CDK14 expression and other clinical variables, such as age or sex. Spearman’s correlation tests were performed to further explore the correlation between CDK14 and Ki-67 expression by percentage of positive cells in the tumor tissues showed a positive correlation in ESCC (P=0.012; Figure 2A).
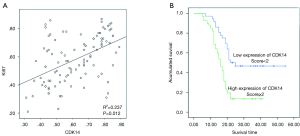
These data prompted us to investigate if CDK14 could be an independent prognostic factor of ESCC. Pearson’s χ2 tests were conducted for survival analyses. CDK14 expression significantly correlated with survival status (P=0.000) (Table 2). Specifically, 17 of 36 (47.2%) patients in the group with low CDK14 expression survived compared to 9 of 67 (13.4%) in the group with high CDK14 expression. Kaplan-Meier analysis showed that high CDK14 expression was significantly related to poor survival (P=0.000; Figure 2B). These data showed that CDK14 was an independent prognostic indicator for ESCC patients.
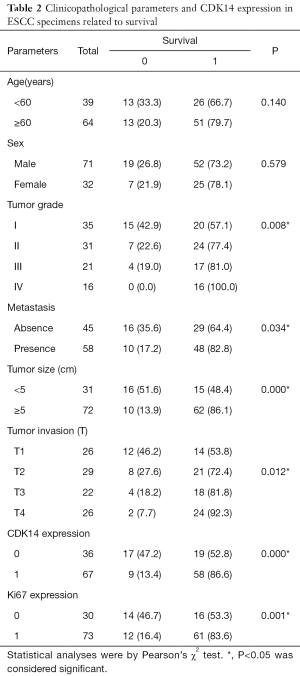
Full table
CDK14 expression and cell cycle development in ESCC
Research shows that CDK14 promotes cell cycle progression in various cancers. Thus, we hypothesized that CDK14 expression may also regulated progression of the cell cycle in ESCC cells. CDK14 expression was higher in Eca109 cells at 12 h after serum stimulation than in cells starved without serum for 72 h (Figure 3A). Constitutive expression of caspase-3 suggested that decreased CDK14 expression was not due to apoptosis. We investigated CDK14 expression by proliferation status of ESCC cells. We found that 66.8% of ECA109 cells arrested in G1 phase after serum starvation for 72 h (S72). Upon refeeding with 10% FBS-containing medium, cells were released from G1 phase and gradually reentered S phase (Figure 3B,C). Western blots showed that CDK14 expression increased as early as 3 h after serum stimulation (R3h) and reached its highest level after serum re-addition (R9h) (Figure 3D). CDK14 expression aligned with cyclin E expression, which is a cell proliferation maker with expression mainly at the G1/S phase (17), as well as the expression pattern of PCNA in different cell cycle phases. This result suggested that CDK14 might function as a positive regulator of ESCC cells in a cell cycle-dependent manner.
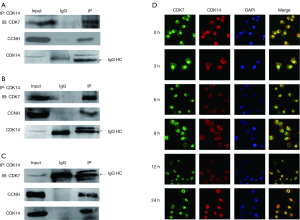
CDK14 interacted with CDK7/CCNH in tissue and ESCC cells
Protein spectrum analysis suggested that CDK14 interacted with CDK7/CCNH, which suggested exploring the mechanism of CDK14 involvement in ESCC. Co-immunoprecipitation experiments were conducted on ESCC tissues and Eca109 cell lysates using anti-CDK14 or anti-CDK7 (Figure 4A,B,C). CDK14 co-immunoprecipitated specifically with anti-CDK7, but not in control conditions. This result demonstrated a naturally occurring interaction between endogenous CDK14 and CDK7 in vivo and vice versa. Endogenous interaction between CDK14 and CCNH was verified in tissues and ESCC cells. Immunofluorescence assays determined the cellular localization of interaction in ESCC cells with different proliferating status. This result showed that CDK14 had a distribution in the cytoplasm or nucleus and CDK14 had physiological interactions with CDK7 in the cell nucleus of Eca109 cells (Figure 4D). But the mechanism for PFTK1 shuttling between the cytoplasm and nucleus need to be further investigated in the future research.
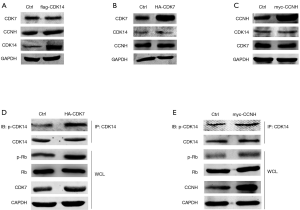
CDK7/CCNH complex activated CDK14 by phosphorylation which phosphorylated the substrate Rb
Above data showed that CDK14 interacted with CDK7 in the nucleus of Eca109 cells. Previous studies showed that CDK7 is a catalytic subunit in the CAK complex that activates CDKs including CDK2, CDK4 and CDK6 by phosphorylation, which promotes cell cycle progression (18). Therefore, we examined how CDK14 interacted with CDK7 and if the interaction of CDK14/CDK7 activated CDK14 in the ESCC cell line. Eca109 cells were serum-starved for 24 h in preparation for synchronization. Cells were transiently transfected respectively with CDK7 and CCNH plasmids to explore the effect of overexpression. No protein level changes of CDK14 were observed (Figure 5A,B,C). Based on co-immunoprecipitation analysis with anti-CDK14 and western blots with anti-Thr/Ser, CDK7 overexpression increased phosphorylation levels of CDK14 and phosphorylated retinoblastoma (p-Rb). Total protein levels did not change (Figure 5D). Rb, a cancer suppressor, deactivates when phosphorylated (19). As a consequence, E2F separates from Rb to regulate the expression of genes essential for a range of functions including cell cycle progression (20). The effect of overexpression of CCNH on CDK14 activity was not obvious, which could be explained by CCNH being a regulated unit in the CDK7/CCNH complex (Figure 5E) (21). The results indicated that the mechanism of CDK14 involvement in ESCC proliferation was activated by phosphorylation via crosstalk with CDK7/CCNH in ESCC cells.
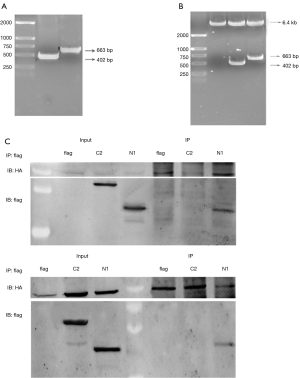
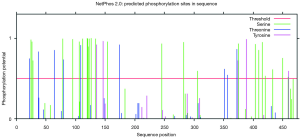
We constructed two truncated mutants of CDK14: N1 (1-134 amino acid, aa) and C2 (230-451 aa) (Figure S1A,B). The N1 mutant but not the C2 mutant co-immunoprecipitated with CDK7. The same result was observed in co-immunoprecipitation assays with CCNH (Figure S1C). Amino acid sequence analysis of CDK14 by bioinformatics software suggested a number of threonine and serine phosphorylation sites in the domain 1-134 aa (Figure S2). This result supported the conclusion that CDK14 was activated by the CDK7/CCNH complex, which phosphorylated Rb to inactivate it.
Upregulation of CDK14 promoted proliferation and enhanced cisplatin resistance in ESCC
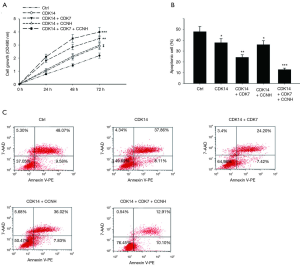
We investigated the role of CDK14 in ESCC proliferation. To explore if CDK14 high expression caused Eca109 cell proliferation, colony formation experiments were performed with cells transfected for 48 h. Colony formation was significantly higher with CDK14 overexpression than with control conditions. Co-overexpression of CDK7 and CDK14 enhanced this effect on Eca109 cells but not when CCNH was co-transfected with CDK14 (Figure 6A). Cell proliferation was measured and showed similar results (Figure 6B). Because high CDK14 expression was related to poor response to chemotherapy in ESCC, we further explored the resistance of the ESCC cell line to cisplatin. Cisplatin is a first-line treatment for several human cancers including esophageal cancer (22). Our previous study revealed that the IC50 of cisplatin for the Eca109 cell line is 40 nM with 48 h treatment (23). That concentration of cisplatin was used in the following experiments. Annexin V-FITC/PI assays showed a significant decrease in cell death with increased expression of CDK14 when Eca109 cells were treated with cisplatin compared with a control group. These data suggested that CDK14 enhanced resistance to cisplatin in ESCC. Co-overexpression of CDK7 and CDK14 enhanced this effect on Eca109 cells but not when CCNH was co-transfected with CDK14 (Figure 6C). When three plasmids which were CDK14, CDK7 and CCNH were co-transfected, chemo-sensitivity to cisplatin decreased. Our results showed that CDK14 promoted cell proliferation and one activation mechanism of CDK14 providing more potential treatment targets for ESCC patients.
Discussion
The evolution and development of esophageal cancer as well as chemoresistance are closely related to cell cycle disorder (24,25). Unregulated cell growth and proliferation is the main feature of tumors. Cell cycle proteins (cyclin), CDKs and cyclin-dependent kinase inhibitor (CKI) are involved in the regulation of the cell cycle. Factors that promote tumor occurrence such as cyclin D1, cyclin B, CDK2, E2 or inhibit tumor occurrence such as p16, p21, p53, and Rb also have key functions in the carcinogenesis of the esophagus. Disorder of cell cycle control is important for the occurrence and development of esophageal cancer, and provides a direction for clinical treatment of esophageal cancer and improving patient survival. Mechanism studies of cell cycle regulation in the development of esophageal cancer are important to find effective indexes for clinical diagnosis and treatment.
Throughout the cell cycle which is a continuous process, CDK is a core component, with positive regulation of cyclins and negative regulation of CKIs. Abnormal regulation of CDK is closely related to non-procedural proliferation, genomic instability and chromosomal instability (26). A study found that a variety of CDK disorders are involved in the occurrence and development of esophageal cancer. Increased expression of CDK2 in esophageal cancer is related to tumor progression and CDK4/CDK6 are targets in esophageal cancer treatment (27). Kinase cyclin-dependent 14, CDK14, is a cell cycle regulatory protein that has been intensively studied. CDK14 is highly expressed in esophageal cancer and is associated with resistance to chemotherapy, indicating poor prognosis. It is a novel CDK and its gene is on human chromosome 17q21.13. The protein sequences for the catalytic domain have high homology with the cdc2-related protein family, which regulate cell G1/S and G2/M phase (28). Research on CDK14 found that it is a potential tumor-related gene. Expression of CDK14 is increased in many kinds of tumors, such as in liver and breast cancer. Studies of esophageal cancer show that high CDK14 expression is related to resistance to chemotherapy drugs such as cisplatin, but the mechanism of the cell regulation pathway of CDK14 was unclear.
Our study verified that CDK14 was overexpressed in ESCC tissue compared to normal tissue using western blots. IHC staining revealed that CDK14 was positively correlated with clinical pathological variables such as tumor size, tumor grade and tumor invasion, and Ki-67. Serum starvation and refeeding assays showed that CDK14 expression was positively related to cyclin D1 expression. These results suggested that CDK14 may have a key role in development of esophageal cancer. The results led us to further study the molecular mechanisms of CDK14. Protein spectrum analysis suggested that CDK14 interacted with CDK7/CCNH, a CAK (CDK-activating kinase) that is implicated in activating several types of CDKs including CDK2 and CDK4 by phosphorylation. Our previous work showed that CDK7 and CCNH are highly expressed and advance development of ESCC. Thus, CDK7/CCNH may interact with CDK14 to be a tumor promoter in esophageal cancer. Thus, the function of CDK14 was regulated by a CAK complex of CDK7/CCNH. We demonstrated that CDK14 interacted with CDK7/CCNH in cells and tissues of esophageal cancer. In Eca109 cells, CDK7 had a major role in CDK14 activation but not its protein expression level. The effect of CCNH on CDK14 activation was not obvious. Immunofluorescence showed that the distribution of CDK14 changed from cytoplasm to nucleus after serum refeeding for 3, 6, 9, 12, or 24 h. CDK14 co-localized with CDK7 in the nucleus, suggesting that the CDK7/CCNH complex may activate CDK14 in the nucleus. Many proteins with nuclear localization signals (NLSs) can be imported into the nucleus, mediated by the importin α/b complex. A protein that shuttles between the nucleus and cytoplasm usually contains an NLS that binds to an importin and an NES that binds to an exportin. Two predicted NLSs were located at residues 66 to 72 (PEDKKVR) and 113 to 119 (PKVRRHS) of PFTK1. However, the mechanism for PFTK1 shuttling between the cytoplasm and nucleus need to be further investigated.
CDK14 could then activate downstream signal molecules to affect cell cycle progression. CDK14 clearly upregulated the phosphorylation level of its downstream target, Rb, to deactivate it. Rb is a classic cancer suppressor that promotes the process of the G1/S phase in the cell cycle. Furthermore, we found that upregulation of CDK14 promoted Rb phosphorylation in ESCC. We concluded that in ESCC, CDK14 promoted cell proliferation by upregulating Rb phosphorylation and was activated by a CAK complex of CDK7/CCNH. In vitro, we verified the effect on accelerating the cell cycle at G1/S, promoting cell proliferation and increasing tolerance to cisplatin in the ESCC cell line Eca109 in the CDK14 cell signaling regulation pathway. Cisplatin is the first-line drug for esophageal cancer chemotherapy, and cell sensitivity to cisplatin differs at different periods. The main effect of cisplatin on G1-phase cells was significantly inhibited by cisplatin and the inhibiting effect of cisplatin on S phase was significantly reduced in the Eca109 ESCC cell line by the CDK14 cell signaling regulation pathway.
To analyze the protein domain of CDK14 that was regulated by the CDK7/CCNH complex, we constructed two fragments: N1 (1-134 aa) and C2 (230-451 aa). Full-length CDK14 and the N1 mutant but not the C2 mutant co-immunoprecipitated with CDK7. The same result was observed in co-immunoprecipitation assays with CCNH. Analysis of the amino acid sequence of CDK14 by bioinformatics software suggested a number of threonine and serine phosphorylation sites in the domain 1-134 aa. Fang Su et al. verified that CDK14 has a conserved domain in 134-230 aa, which is present in others in the CDK family and interacts with cyclin D3 and p21. Thus, we cannot exclude that CCNH may directly interact with CDK14 in the conserved domain 134-230 aa. The interaction between CDK14 and CCNH might be indirect by crosstalk between CDK7 and CDK14, requiring further experimentation to confirm. However, CDK14 can be phosphorylated and further activated, mainly by CDK7 at the N1 amino acid domain. In vivo, the experiments showed that the N1 domain promoted proliferation and decreased apoptosis after treatment of Eca109 cells with cisplatin.
We demonstrated CDK14 involvement in the malignant development of tumor cells by affecting cell proliferation, which could lead to drug resistance. The mechanism was CDK14 activation by a CAK complex of CDK7/CCNH, causing Rb inactivation by phosphorylation in the nucleus. The results provided new evidence about CDK14 in the development of esophageal cancer and provide a basis for clinical treatment.
Acknowledgments
Funding: This work was supported by grants from the Nantong Science and Technology Project to SJ Ni (MS22016068), and the Natural Science Foundation of Nantong University (14Z013).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by Ethics the Committee of Affiliated Hospital of Nantong University (ID: 2013-67). Informed consent was provided by all participants.
References
- Li B, Li J, Xu WW, et al. Suppression of esophageal tumor growth and chemoresistance by directly targeting the PI3K/AKT pathway. Oncotarget 2014;5:11576-87. [PubMed]
- Bray F, Ren JS, Masuyer E, et al. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer 2013;132:1133-45. [Crossref] [PubMed]
- McGuire S.. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr 2016;7:418-9. [Crossref] [PubMed]
- Wu C, Wang Z, Song X, et al. Joint analysis of three genome-wide association studies of esophageal squamous cell carcinoma in Chinese populations. Nat Genet 2014;46:1001-6. [Crossref] [PubMed]
- Schmitz ML, Kracht M. Cyclin-Dependent Kinases as Coregulators of Inflammatory Gene Expression. Trends Pharmacol Sci 2016;37:101-13. [Crossref] [PubMed]
- Asghar U, Witkiewicz AK, Turner NC, et al. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov 2015;14:130-46. [Crossref] [PubMed]
- Roskoski R Jr. Cyclin-dependent protein kinase inhibitors including palbociclib as anticancer drugs. Pharmacol Res 2016;107:249-75. [Crossref] [PubMed]
- Yang T, Chen JY. Identification and cellular localization of human PFTAIRE1. Gene 2001;267:165-72. [Crossref] [PubMed]
- Gao Y, Jiang M, Yang T, et al. A Cdc2-related protein kinase hPFTAIRE1 from human brain interacting with 14-3-3 proteins. Cell Res 2006;16:539-47. [Crossref] [PubMed]
- Pang EY, Bai AH, To KF, et al. Identification of PFTAIRE protein kinase 1, a novel cell division cycle-2 related gene, in the motile phenotype of hepatocellular carcinoma cells. Hepatology 2007;46:436-45. [Crossref] [PubMed]
- Gu X, Wang Y, Wang H, et al. Upregulated PFTK1 promotes tumor cell proliferation, migration, and invasion in breast cancer. Med Oncol 2015;32:195. [Crossref] [PubMed]
- Sun T, Co NN, Wong N. PFTK1 interacts with cyclin Y to activate non-canonical Wnt signaling in hepatocellular carcinoma. Biochem Biophys Res Commun 2014;449:163-8. [Crossref] [PubMed]
- Miyagaki H, Yamasaki M, Miyata H, et al. Overexpression of PFTK1 predicts resistance to chemotherapy in patients with oesophageal squamous cell carcinoma. Br J Cancer 2012;106:947-54. [Crossref] [PubMed]
- Wan C, Gong C, Zhang H, et al. Beta2-adrenergic receptor signaling promotes pancreatic ductal adenocarcinoma (PDAC) progression through facilitating PCBP2-dependent c-myc expression. Cancer Lett 2016;373:67-76. [Crossref] [PubMed]
- Wang Y, Wang Y, Xiang J, et al. Knockdown of CRM1 inhibits the nuclear export of p27(Kip1) phosphorylated at serine 10 and plays a role in the pathogenesis of epithelial ovarian cancer. Cancer Lett 2014;343:6-13. [Crossref] [PubMed]
- Zhang J, Yang X, Wang Y, et al. Low expression of cyclinH and cyclin-dependent kinase 7 can decrease the proliferation of human esophageal squamous cell carcinoma. Dig Dis Sci 2013;58:2028-37. [Crossref] [PubMed]
- Scaglione-Sewell BA, Bissonnette M, Skarosi S, et al. A Vitamin D3 Analog induces a G1-phase arrest in CaCo-2 cells by inhibiting Cdk2 and Cdk6: Roles of cyclin E, p21Waf1, and p27Kip1. Endocrinology 2000;141:3931-9. [Crossref] [PubMed]
- Bisteau X, Paternot S, Colleoni B, et al. CDK4 T172 phosphorylation is central in a CDK7-dependent bidirectional CDK4/CDK2 interplay mediated by p21 phosphorylation at the restriction point. PloS Genet 2013;9:e1003546. [Crossref] [PubMed]
- Matsui T, Nieto-Estevez V, Kyrychenko S, et al. RB controls growth, survival, and neuronal migration in human cerebral organoids. Development 2017;144:1025-34. [Crossref] [PubMed]
- Wu N, Jia D, Bates B, et al. A mouse model of MYCN-driven retinoblastoma reveals MYCN-independent tumor reemergence. J Clin Invest 2017;127:888-98. [Crossref] [PubMed]
- Wang Y, Liu F, Mao F, et al. Interaction with cyclin H/cyclin-dependent kinase 7 (CCNH/CDK7) stabilizes C-terminal binding protein 2 (CtBP2) and promotes cancer cell migration. J Biol Chem 2013;288:9028-34. [Crossref] [PubMed]
- Wang HY, Yao ZH, Tang H, et al. A retrospective clinical study of comparing paclitaxel plus S-1 versus paclitaxel plus cisplatin as the first-line treatment for patients with advanced esophageal squamous cell carcinoma. Oncotarget 2017;8:7540-7. [PubMed]
- Chen L, Ni S, Li M, et al. High expression of BCCIP beta can promote proliferation of esophageal squamous cell carcinoma. Dig Dis Sci 2017;62:387-95. [Crossref] [PubMed]
- Xu R, Wang F, Wu L, et al. A systematic review of hypermethylation of p16 gene in esophageal cancer. Cancer Biomark 2013;13:215-26. [Crossref] [PubMed]
- Michaelsen SH, Larsen CG, von Buchwald C. Human papillomavirus shows highly variable prevalence in esophageal squamous cell carcinoma and no significant correlation to p16INK4a overexpression: a systematic review. J Thorac Oncol 2014;9:865-71. [Crossref] [PubMed]
- Nabel EG. CDKs and CKIs: molecular targets for tissue remodelling. Nat Rev Drug Discov 2002;1:587-98. [Crossref] [PubMed]
- Matsumoto M, Furihata M, Ishikawa T, et al. Comparison of deregulated expression of cyclin D1 and cyclin E with that of cyclin-dependent kinase 4 (CDK4) and CDK2 in human oesophageal squamous cell carcinoma. Br J Cancer 1999;80:256-61. [Crossref] [PubMed]
- Platholi J, Federman A, Detert JA, et al. Regulation of protein phosphatase 1I by Cdc25C-associated kinase 1 (C-TAK1) and PFTAIRE protein kinase. J Biol Chem 2014;289:23893-900. [Crossref] [PubMed]


