
Investigation of the underlying hub genes and mechanisms of reperfusion injury in patients undergoing coronary artery bypass graft surgery by integrated bioinformatic analyses
Introduction
Cardiovascular disease (CVD) has been the leading cause of mortality and morbidity in the past few decades (1). In the past 10 years, the number of people dying from CVD has increased by 12.5% (2). The burden of CVD has become a great challenge worldwide. Among all kinds of CVDs, ischemic heart disease (IHD) accounts for most of the global CVD burden. Coronary artery bypass graft (CABG) surgery is the main method to revascularize the severe multivessel lesion arteries (3). However, the surgery is complicated as a result of the perioperative injury caused by acute cardiovascular ischemia-reperfusion (IR) damage during the process of executing the cardiopulmonary bypass (CPB) (4,5). Also, due to an aging population and the higher ratio of comorbidities (such as diabetes mellitus and hypertension), patients undergoing CABG surgery are more prone to reperfusion injury. Therefore, ways to mitigate the cardio-injury of reperfusion during CABG operation are of great interest for clinical doctors. Although many novel cardioprotective therapies appear to be efficient in animal models (including preconditioning, nitric oxide, hypothermia), the vast majority of these therapies are controversial in the clinical field (6-9). The reasons for the failure to translate the therapeutic approaches of the basic research world into the “real world” are complicated. A systematic investigation into the relevant hub genes and mechanisms of IR in patients undergoing the operation may be helpful to reduce IR injury.
Recently, the system biology analyses of microarrays have emerged as powerful tools for uncovering potentially meaningful genes and possible pathophysiological pathways of multiple diseases (10,11). Compared with traditional gene-gene correlation co-expression matrices, the weighted gene co-expression network analysis (WGCNA) utilizes a new method to find highly correlated genes in microarrays. By introducing a soft-threshold algorithm, WGCNA avoids identifying connected genes by setting a hard threshold (12,13). By clustering genes of similar patterns into the same module, the relationship between modules and phenotypes can be calculated by WGCNA. Because of this, WGCNA has been widely used in the study of a variety of pathologies, including in the areas of cardiovascular and neurodegenerative disease (14-16).
Although various bioinformatics analyses of reperfusion in other organs and animals have been performed, a systematic and thorough analysis of cardiac IR in human hearts is still lacking (17,18). Therefore, in this study, a co-expression correlation network was constructed using the expression data from the gene expression omnibus (GEO) database. Two modules, green and yellow, were significantly correlated with IR injury. Enrichment analyses of gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway were executed to characterize the identified differentially expressed genes (DEGs) and selected modules. Also, genes with high intramodular connectivity (IC) in the modules were assessed to identify hub genes in these two modules. Possible transcription factors (TFs) and microRNAs (miRNAs), which might regulate genes in the significant modules, were predicted using the corresponding bioinformatics tools. We concluded that during the IR process in the human atrial samples of patients undergoing CABG surgery, the inflammation response and complement cascade regulation might play pivotal roles. IL-6, CXCL8, IL-1β, MYC, and PTGS-2 were important upregulated genes, while C3, TIMP1, VSIG4, SERPING1, CD163, and HP were key downregulated hub genes. The prediction of potential miRNAs and TFs revealed that miRNAs (hsa-miR-333-5p, hsa-miR-26b-5p, hsa-miR-124-3p, hsa-miR-16-5p, hsa-miR-98-5p, hsa-miR-17-5p, hsa-miR-93-5p) and TF (STAT1) might regulate the gene expression in the most positively related module, while hsa-miR-333-5p and HSF-1 were predicted to regulate the genes within the most negatively related module.
Methods
Medical ethics
The raw dataset was available from the GEO database (http://www.ncbi.nlm.nih.gov/geo/; GSE29396).
In our study, neither human trials nor animal experiments were executed.
Data collection and processing
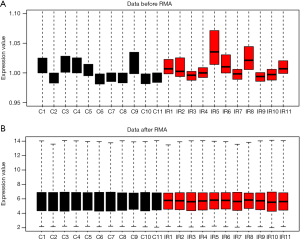
The RNA expression profiles of atrial appendages in patients undergoing CABG surgery were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/; GSE29396). The series was performed on the GPL5175 platform of the Affymetrix Human Exon 1.0 ST Array chip (Thermo Fisher Scientific, MA, USA). The GEO series contains 11 atrial samples collected at the time of cannulation and 11 atrial samples obtained at 15 min after releasing the cross camp during the surgery. Considering that this study aimed to explore the potential pathophysiological changes during the IR process in patients undergoing the operation, atrial samples from patients pretreated with remote ischemic preconditioning were excluded. The raw data of the CEL files were read using the oligo package in Bioconductor and subsequently processed using the robust multiarray average (RMA) algorithm (Figure 1) (19). After that, the gene symbols of probes were annotated using the annotation profiles provided by Thermo Fisher. For multiple probes matched to one gene symbol, the mean expression of probes was calculated as the final value of the given gene. The DEGs were identified using linear models for microarray data (limma) in the R package (20). The cut-off criteria were set as a P value <0.05 and | log2-fold change (log2 FC) | ≥0.5.
Enrichment analyses of GO and KEGG pathway
Functional enrichment analyses of selected genes were calculated using clusterProfiler package (20). A P value <0.05 was considered to be significant, and the identified significant analyses were sorted by gene counts.
Protein-protein interaction (PPI) network construction
The PPI network was constructed with a threshold of medium confidence ≥0.4 by the search tool for the retrieval of interacting genes (STRING) database. Then, the network was visualized in Cytoscape software (v3.6.1; http://www.cytoscape.org/).
Correlation of the co-expression network
Instead of constructing a co-expression network of all DEGs, we performed WGCNA analyses in all genes identified in GSE29396 for a better understanding of the co-expression relations within genes. An appropriate soft-threshold was selected to ensure a scale-free topology (R2 >0.85). The topological overlap matrix (TOM) was constructed to measure the network connectivity of the genes. Average linkage hierarchical clustering was performed to cluster genes with similar patterns into the same modules with a minimum size of 30. The relationships between phenotypes and modules were calculated to identify the highly related modules. Finally, highly correlated modules were further analyzed to explore the potential roles of these modules.
Construction of potential TF- and miRNA-target regulatory networks
The iRegulon plugin in Cytoscape was used to predict potential TFs in regulating genes in the highly related modules (21). The predicted TFs with the highest normalized enrichment score (NES) were selected to construct TF-target networks. miRNA-target networks in the highly related modules were built by miRNet analyses (22). All predicted TF- and miRNA-target networks were visualized in Cytoscape.
Results
DEG identification and hierarchical clustering analysis
The raw data were processed by the oligo package, and a matrix of 21,900 probes was identified. The probes were reannotated with the latest annotation files downloaded from Affy. After RMA modification, the median expression value of each gene was calculated. An expression profile of 15,671 genes was obtained.
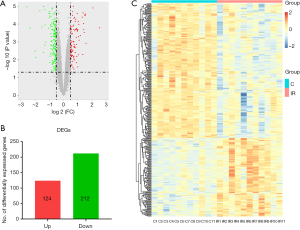
A total of 336 DEGs were identified between the control group and the IR group, including 124 upregulated genes and 212 downregulated genes [P value <0.05, | (log2 FC) | ≥0.5, Figure 2A,B]. Also, hierarchical clustering analysis was also performed for all DEGs between the two groups (Figure 2C). The general expression of the DEGs was significantly different.
Enrichment analyses of all DEGs
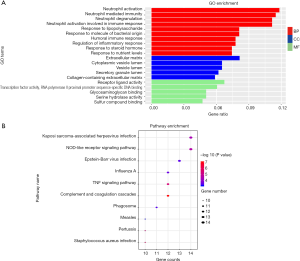
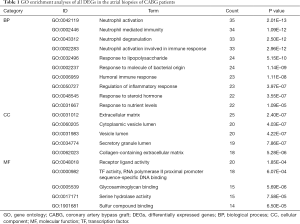
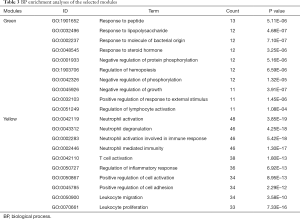
Functional enrichment analyses of biological process (BP), cellular component (CC), and molecular function (MF) were executed by the clusterProfiler package to reveal GO categories of reperfusion injury around CABG surgery. The most enriched BP terms of all DEGs were mainly related to neutrophil activation, neutrophil-mediated immunity, neutrophil degranulation, neutrophil activation involved in immune response, and response to lipopolysaccharide. For the CC categories, major DEGs were mainly associated with the extracellular matrix, cytoplasmic vesicle lumen, vesicle lumen, and secretory granule lumen. Additionally, DEGs were mainly enriched in receptor-ligand activity, TF activity, RNA polymerase II proximal promoter sequence-specific DNA binding (Figure 3A). Detailed information on the GO analyses is listed in Table 1.
Full table
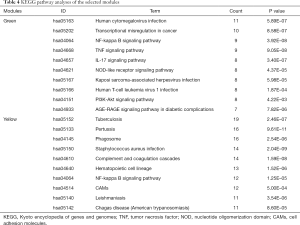
The potential pathways were predicted in the R language (P value <0.05) to screen out the most significant KEGG pathways in IR injury. KEGG pathways were mainly associated with the nucleotide oligomerization domain (NOD)-like receptor signaling pathway, tumor necrosis factor (TNF) signaling pathway, and complement and coagulation cascades (Figure 3B, Table 2).

Full table
Construction of the co-expression network and key module identification
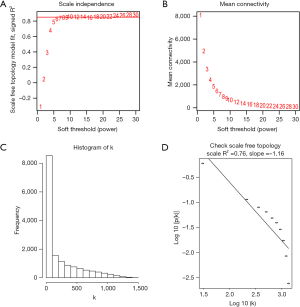
In this study, we constructed the co-expression network by WGCNA to investigate the phenotypic changes of genes associated with IR injury. The 22 samples were clustered using flashClust tools in the WGCNA package to detect outliers (cutHeight =70, Figure S1). Therefore, the expression profile of IR5 and the corresponding profile of C5 were excluded from the following analyses. Before constructing the weighted co-expression matrix, a suitable soft-threshold β was calculated to ensure a scale-free topology. When the soft-threshold β=10, the independence degree went up to 0.8520. The co-expression gene modules were constructed by setting β=10 in the following studies (Figure 4).

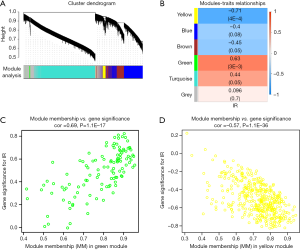
After that, a co-expression network of 15,672 genes was constructed using a one-step method. The correlation matrix was built to calculate the correlation efficiency between genes. Genes with similar expression patterns were clustered into one module. In both the control and IR groups, a total of five modules were identified by the average linkage clustering algorithm (Figure 5A). The module eigengenes (MEs) of the green module (r=0.63, P=3E–3) and yellow module (r=–0.71, P=4E–4) were calculated (Figure 5B). These two modules were identified as the two significant modules with the highest correlations with IR. Therefore, we chose these modules of interest for use in the subsequent analyses. The values between module membership (MM) and gene significance (GS) were calculated by a labeled heatmap function to confirm the correlation between IR and modules of interest. Both the green (cor =0.69, P=1.1E–17) and yellow (cor =–0.57, P=1.1E–36) modules showed high correlations with IR injury (Figure 5C,D).
Functional annotation of the gene modules of interest
As the green and yellow modules were identified as modules of interest, functional annotation of the selected modules was adopted to characterize these modules. Enrichment analyses of GO and KEGG pathways were executed using the clusterProfiler method. The green module was positively correlated with IR injury in operation. On the one hand, by analyzing the BP of the green module, the genes in this module were mainly enriched in response to peptide, lipopolysaccharide, and molecules of bacterial origin, along with the negative regulation of protein phosphorylation, and the positive regulation of response to external stimuli (Table 3). On the other hand, the genes of the yellow module were mainly concentrated in neutrophil activation, neutrophil-mediated immunity, T cell activation, regulation of the inflammatory response, and leukocyte migration (Table 3).
Full table
Additionally, the enriched pathways of the selected modules were investigated. The enriched pathways of the green module were mainly associated with the NF-kappa B signaling pathway, TNF signaling pathway, IL-17 signaling pathway, NOD-like receptor signaling pathway, and AGE-RAGE signaling pathway in diabetic complications (Table 4). For the yellow module, the genes were mainly enriched in complement and coagulation cascades, the NF-kappa B signaling pathway, and cell adhesion molecules (CAMs) (Table 4).
Full table
PPI network construction and identification of hub genes
The IC of each gene was calculated by summing the connection strengths with other genes in the same module. For a given gene, the higher the IC is, the stronger its relations with other genes. The top 100 genes of the green and yellow modules ranked by IC were selected to construct PPI networks by setting medium confidence (0.400) in STRING. Also, hub genes were identified according to the degrees in the PPI networks. In the PPI network of the green module, interleukin 6 (IL-6, degree 36), C-X-C motif chemokine ligand 8 (CXCL8, degree 30), interleukin 1 beta (IL-1β, degree 27), MYC proto-oncogene, bHLH TF (MYC, degree 25), and prostaglandin-endoperoxide synthase 2 (PTGS-2, degree 24) were identified as hub genes with the top 5 highest degrees (Figure 6). For the yellow module, complement C3 (C3, degree 14), TIMP metallopeptidase inhibitor 1 (TIMP1, degree 9), V-set and immunoglobulin domain containing 4 (VSIG4, degree 9), serpin family G member 1 (SERPING1, degree 9), CD163 molecule (CD163, degree 8), and haptoglobin (HP, degree 8) were identified as the hub genes with the highest degrees (Figure 7). In the yellow module, both CD163 and HP have 8 degrees. Above all, these 11 genes were identified as hub genes (Table 5).



Full table
Prediction of potential miRNA-target regulatory networks
In addition to the above, miRNAs, a group of noncoding RNAs that function as important posttranslational regulators in many pathophysiological processes, were also found to play an important role in reperfusion injury (23,24). In this study, we predicted the potential regulatory miRNAs that regulate the top 100 genes of the green and yellow modules ranked by IC using miRNet, a tool which integrates data from 11 different miRNA databases (22). The potential miRNA-target networks were visualized by the miRNet database. A total of 1,582 miRNAs were predicted in the green module. Among these miRNAs, 7 miRNAs (hsa-miR-333-5p, hsa-miR-26b-5p, hsa-miR-124-3p, hsa-miR-16-5p, hsa-miR-98-5p, hsa-miR-17-5p, hsa-miR-93-5p) had degrees ≥15 (Figure 8A). For the yellow module, a total of 1,004 miRNAs were predicted, and hsa-miR-333-5p was predicted to be a potential miRNA (degree 15, Figure 8B).
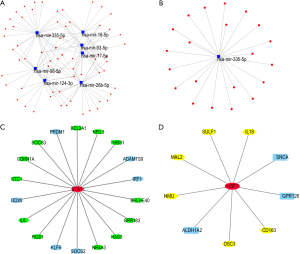
TF network construction
TFs are proteins that bind to specific DNA sequences to control the rate of transcription information from DNA to mRNA. Here, we predicted the potential TFs in the module of interest using the iRegulon plugin in Cytoscape software. For each module, the predicted TF with the highest NES was displayed. In the green module, STAT1 (NES 6.549) was predicted to regulate 18 targets (BCL2A1, THBS1, ADAMTS9, BHLHE40, GPR183, HAS2, NR4A3, SOCS2, KLF6, RGS1, IL-6, DDX5, STC1, CDKN1A, SOCS3, PRDM1, NFIL3, IRF1) (Figure 8C). In the yellow module, HSF-1 (NES 5.986) was predicted to be the potential TF with the highest NES to regulate 9 targets (CD163, DSC3, ALDH1A2, NMU, MAL2, SULF1, IL-18, SNCA, GPR126) (Figure 8D).
Discussion
Despite an array of fundamental research devoted to studying the reduction of intraoperative IR injury, its translation into a clinically relevant protective strategy remains elusive (25). The huge heterogeneity between animals and humans may play an important role in the difficulty of translation (26). In this study, a systematic, integrated bioinformatic approach was adopted to explore the potential hub genes and important pathways underlying IR injury in the heart tissues collected in patients undergoing CABG surgery. It was found that IL-6, CXCL8, IL-1β, MYC, and PTGS-2 might be hub upregulated genes, while C3, TIMP1, VSIG4, SERPING1, CD163, and HP might be hub downregulated genes. To our knowledge, this is the first systematic bioinformatics study to analyze the gene profiles of IR injury in human heart samples.
In our study, we identified 336 DEGs that were significantly differentially expressed in the human atrial samples collected before the time of revascularization and after releasing the cross-clamp. Enrichment analyses of GO were used to characterize these DEGs, and the most enriched GO terms of BP were mainly associated with neutrophil activation, neutrophil-mediated immunity, neutrophil degranulation, neutrophil activation involved in the immune response, and regulation of the inflammatory response. In the CC terms, most DEGs were enriched in the extracellular matrix, cytoplasmic vesicle lumen, and secretory granule lumen. In the MF category, GO terms were mainly associated with receptor-ligand activity and serine hydrolase activity. KEGG analyses of these DEGs revealed that the NOD-like receptor signaling pathway, TNF signaling pathway, and complement and coagulation cascades might be related to changes in gene expression profiles.
Through the enrichment of 336 DEGs, we determined that neutrophils might play a pivotal role in mediating IR injury. The concepts of “neutrophil activation” and “regulation of inflammatory response” might fit well with the inflammatory feature of acute IR injury in the heart as described previously (27). Indeed, CABG surgery has been shown to provoke the inflammatory response through the restoration of perfusion by releasing the aortic cross-clamp (28). This process is concomitantly related to the release of cytokines, such as IL-1β, IL-6, TNF-α, and CXCL8 (29-31). Interestingly, IL-6, IL-1β, CXCL8 were also predicted to upregulate hub genes in our study. TNF-α was also found to be significantly upregulated (data not shown). Meanwhile, KEGG analyses indicated the role of the NOD-like receptor signaling pathway, TNF signaling pathway, and complement and coagulation cascades, which is also following previous reports (32,33).
Instead of constructing a correlation network of all DEGs, a weighted gene co-expression network based on all annotated genes was constructed. A total of five modules were recognized, and the green and yellow modules were the highest positively and negatively correlated modules with IR injury. Most genes enriched in the green modules were associated with response to peptide, response to lipopolysaccharide, and positive regulation of response to external stimuli. On the other hand, the yellow module was mainly enriched in immune responses. According to the predicted results of the module genes, we determined that the process of reperfusion initiated the inflammatory response and activated a series of chemokines. Also, instead of identifying hub genes through the PPI network of all DEGs, our study identified hub genes through the related modules from WGCNA. Five upregulated hub genes (IL-6, CXCL8, IL-1β, MYC, PTGS-2) and 6 downregulated hub genes (C3, TIMP1, VSIG4, SERPING1, CD163, HP) were screened from the two selected modules.
Among the 5 upregulated hub genes, IL-6 was the hub gene with both the biggest log2 FC and the highest degree. IL-6 was a pleiotropic cytokine with an original immunity role in response to acute infection (34-36). Previous studies have shown that IL-6 is elevated in cardiac surgery with CPB (37). Moreover, gene polymorphisms of IL-6 (–572G > C and –174G > C) have also been related to prolonged hospitalization after cardiac surgery (38). The level of IL-6 was related to the severity of the cardiac injury, with high IL-6 levels indicating the reduced left ventricular ejection fraction (LVEF) in patients diagnosed with acute coronary syndrome (ACS) (39,40). Another study showed that a single intraperitoneal injection of the anti-IL-6 receptor antibody (MR 16-1) to inhibit the IL-6 receptor in mice subjected to left anterior descending (LAD) coronary artery ligation significantly ameliorated cardiac inflammation and prevented left ventricular remodeling (41). However, the administration of MR 16-1 failed to obtain the protective effect of cardiac remodeling in mice undergoing IR injury (LAD ligation for 45 min) (42). In our study, we found that IL-6 was expressed 5.81-fold higher in the atrial samples before perfusion than in the heart samples after perfusion. However, whether targeting IL-6 would be beneficial for the recovery of cardiac function in patients undergoing CABG remains to be explored in subsequent clinical studies. For other predicted upregulated genes, CXCL8 is a CXC chemokine that has been shown to activate both CXCR1 and CXCR2 to recruit neutrophils (43). CXCL8 was found to be upregulated in the infarcted myocardium tissue in the animal models (44). Previous studies have proven that the suppression of CXCL8 ameliorated IR injury in the retina of rats (45). However, CXCL8 was found to exhibit a protective effect in a rabbit IR model (46). Similar to CXCL8, the role of IL-1β remains to be explored in further experiments. Usually, IL-1β is elevated in the reperfusion tissue, and high levels of IL-1β have been associated with poor prognosis (47-49). However, targeting IL-1β may be beneficial for the cardiac function of patients suffering from IR injury.
Moreover, MYC, originally recognized as an important oncogene that regulates the cell cycle, was recently found to be involved in the progression of CVD (50). In the cerebral IR models, increased reactive oxygen species (ROS) regulated MYC activity, and blockade of MYC seemed to reduce IR injury (51). Upregulated MYC was related to accelerated fibroblast proliferation after IR injury; therefore, suppressing MYC might ameliorate adverse remodeling after IR injury (52). PTGS-2, also known as cyclooxygenase (COX)-2, was increased significantly in response to various stimuli, including IR injury (53,54). The suppression of PTGS-2 was found to alleviate IR injury to some degree. Aspirin, a regularly used antiplatelet drug, was also a nonselective COX inhibitor. Therefore, the role of PTGS-2 in clinical studies might be disrupted by the use of aspirin (55).
In contrast to previous studies, the gene expression value of C3 was significantly downregulated in the atrial samples after reperfusion (56). C3 was a major component of the complement system. During IR injury, the complement cascade was activated (56). Due to the relatively short half-life of C3 in vivo, after synthesis, C3 was immediately cleaved into C3b and C3a (57). C3b covalently attached to the target cells and was subsequently degraded into C3dg and C3d. Compared with C3 and C3b, C3b had a longer half-life and could be detected for several days in situ (58). In several IR models in animals, the expression of C3d was associated with the degree of tissue injury (59,60). Therefore, C3d was often used as a marker of the tissues from patients suffering from IR injury. In the samples from patients that died from ACS, the deposition of C3d was higher, regardless of receiving reperfusion or not, than that in in the people without ACS (56).
Furthermore, the suppression of C3 helped reduce cerebral IR injury in mice models (61). However, C3 was significantly downregulated in our analyses. The samples collected after IR injury were obtained at 15 minutes after releasing the cross-clamp. Compared with previous research, the gene expression of C3 in our study was detected very shortly after IR injury. Also, activation of the complement cascade during IR injury was mainly attributed to accelerated C3 cleavage (62). Considering that the goal of this study was to explore the general gene variations in human samples, the role of C3 might be explored in future studies.
Similarly, TIMP1, a specific inhibitor of matrix metalloproteinase-9 (MMP-9), was also found to be significantly elevated 24 hours after IR injury in several animal models (63,64). However, TIMP1 was found to alleviate IR injury in the cerebral IR models (64). Therefore, the expression of TIMP1 might be dynamic and varied according to the duration of IR injury. Another hub gene, VSIG4, was a receptor for the complement component, including C3 (65). Interestingly, a recent study showed that inhibition of VSIG4 enhanced the expression of IL-1β, the predicted upregulated gene (66). Although there are no studies exploring the effect of VSIG4 on IR injury, it is reasonable to assume that the role of VSIG4 in the cardiovascular system can be ascertained in future studies.
Similarly, SERPING1, a C1 inhibitor encoding gene, was also predicted to be downregulated in our study. A previous study reported that SERPING1 inhibited activated C1 components and regulated the complement cascade (67). Emmens et al. found that SERPING1 was downregulated in the first 2 hours in a rat IR model but increased 10-fold after 4 days (68).
Moreover, both CD163 and HP have predicted hub genes with degree 8. CD163 was originally found to be a macrophage-specific receptor for hemoglobin:haptoglobin (HB:HP) complexes (69). The HP-CD163 signaling pathway is a captor-receptor system that activates heme oxygenase-1 (HO-1) (70). Many studies in mice have demonstrated that HO-1 plays an important role in the catabolism of ROS and that defective HO-1 exacerbates IR injury (71). Following the prediction of hub genes, we also predicted the potential miRNAs and TFs in the modules of interest. Many studies have discussed the role of miRNAs and TFs in IR injury (72-75). However, how to translate these studies into clinical application is still uncertain. We hope that our work can indicate potential miRNAs and TFs for future research. Above all, the predicted hub genes suggest that the activation of immune response and the impairment of the complement cascade might play a major role during the super acute phase IR phase in CABG surgery.
The microarray profiles analyzed in this study were originally submitted by Zaugg et al. In our study, we aimed to illustrate the overview of gene expression changes in human atrial samples from patients undergoing CABG surgery using bioinformatics analytical methods. The strategy to reduce cardiovascular IR injury has been a very hot topic in recent decades. Part of the predicted hub genes were also explored in the animal IR models. However, the failure to translate these studies into clinical work inspired us to analyze the overall gene changes by systems biology analysis algorithms.
Furthermore, to reduce the bias in the preliminary study, we constructed the co-expression network by WGCNA, which had been widely used in the microarray profile analyses (76-78). Moreover, considering that our study aimed to identify the potential hub genes and mechanisms underlying the perfusion process in CABG surgery, we selected the modules with the highest MEs related to IR. Subsequently, the enrichment analyses and prediction of potential hub genes were executed within the modules of interest. We did not validate the predicted results in our study because the goal of this study was to illustrate the overall gene expression patterns of IR in patients undergoing CABG surgery within the first few minutes. Indeed, several studies have been devoted to exploring the mechanisms during the reperfusion process in animal models. However, the failure to translate these results into clinical work prompted us to analyze the microarray profiles in human samples. Furthermore, the variations in the hyperacute phase of IR injury were quite dynamic and unstable. The results of validation might be confusing, considering that the exact collection time of atrial samples might be unclear.
In conclusion, our results indicate that during the IR process in the human atrial samples of patients undergoing CABG surgery, the inflammation response and complement cascade regulation might play pivotal roles. IL-6, CXCL8, IL-1β, MYC, and PTGS-2 were important upregulated genes, while C3, TIMP1, VSIG4, SERPING1, CD163, and HP were key downregulated hub genes. Predicted miRNAs (hsa-miR-333-5p, hsa-miR-26b-5p, hsa-miR-124-3p, hsa-miR-16-5p, hsa-miR-98-5p, hsa-miR-17-5p, hsa-miR-93-5p) and TF (STAT1) might regulate gene expression in the most positively related module, while hsa-miR-333-5p and HSF-1 were predicted to regulate the genes within the most negatively related module. Although our results are preliminary, these analyses provided novel insights into the molecular mechanisms of IR injury in human hearts undergoing CABG surgery and might, therefore, facilitate the future translation of this research into clinical work.
Acknowledgments
Funding: This work was supported by a grant from the Natural Science Funds of Zhejiang Province, China (Project No. LY 16H020005, Project No. LY16H020001, Project No. LQ16C110001, Project No. LY16H020008, Project No. LQ17H020001, and Project No. LQ17H020002) and the National Natural Science Foundation of China (81800212 and 81570246). The funders were not involved in the study design, data collection, and analysis, manuscript preparation, or decision to publish.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was not involved in the experiments of humans or animals. The raw dataset was available from the GEO database (http://www.ncbi.nlm.nih.gov/geo/; GSE29396).
References
- Joseph P, Leong D, McKee M, et al. Reducing the global burden of cardiovascular disease, part 1: the epidemiology and risk factors. Circ Res 2017;121:677-94. [Crossref] [PubMed]
- GBD 2015 Mortality and Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016;388:1459-544. [Crossref] [PubMed]
- Hausenloy DJ, Yellon DM. Ischaemic conditioning and reperfusion injury. Nat Rev Cardiol 2016;13:193-209. [Crossref] [PubMed]
- Venugopal V, Ludman A, Yellon DM, et al. 'Conditioning' the heart during surgery. Eur J Cardiothorac Surg 2009;35:977-87. [Crossref] [PubMed]
- Hausenloy DJ, Boston-Griffiths E, Yellon DM. Cardioprotection during cardiac surgery. Cardiovasc Res 2012;94:253-65. [Crossref] [PubMed]
- Lucchinetti E, Bestmann L, Feng J, et al. Remote ischemic preconditioning applied during isoflurane inhalation provides no benefit to the myocardium of patients undergoing on-pump coronary artery bypass graft surgery: lack of synergy or evidence of antagonism in cardioprotection? Anesthesiology 2012;116:296-310. [Crossref] [PubMed]
- Bulluck H, Yellon DM, Hausenloy DJ. Reducing myocardial infarct size: challenges and future opportunities. Heart 2016;102:341-8. [Crossref] [PubMed]
- Hausenloy DJ, Baxter G, Bell R, et al. Translating novel strategies for cardioprotection: the Hatter Workshop Recommendations. Basic Res Cardiol 2010;105:677-86. [Crossref] [PubMed]
- Ludman AJ, Yellon DM, Hausenloy DJ. Cardiac preconditioning for ischaemia: lost in translation. Dis Model Mech 2010;3:35-8. [Crossref] [PubMed]
- Zhang X, Feng H, Li D, et al. Identification of differentially expressed genes induced by aberrant methylation in oral squamous cell carcinomas using integrated bioinformatic analysis. Int J Mol Sci 2018. [Crossref] [PubMed]
- Wang T, Zheng X, Li R, et al. Integrated bioinformatic analysis reveals YWHAB as a novel diagnostic biomarker for idiopathic pulmonary arterial hypertension. J Cell Physiol 2019;234:6449-62. [Crossref] [PubMed]
- Prieto C, Risueno A, Fontanillo C, et al. Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles. PLoS One 2008;3:e3911. [Crossref] [PubMed]
- Chen J, Yu L, Zhang S, et al. Network analysis-based approach for exploring the potential diagnostic biomarkers of acute myocardial infarction. Front Physiol 2016;7:615. [Crossref] [PubMed]
- Wan Q, Tang J, Han Y, et al. Co-expression modules construction by WGCNA and identify potential prognostic markers of uveal melanoma. Exp Eye Res 2018;166:13-20. [Crossref] [PubMed]
- Yin L, Cai Z, Zhu B, et al. Identification of key pathways and genes in the dynamic progression of HCC based on WGCNA. Genes (Basel) 2018;9. [Crossref] [PubMed]
- Liang JW, Fang ZY, Huang Y, et al. Application of weighted gene co-expression network analysis to explore the key genes in Alzheimer's disease. J Alzheimers Dis 2018;65:1353-64. [Crossref] [PubMed]
- Zhu K, Zheng T, Chen X, et al. Bioinformatic analyses of renal ischaemia-reperfusion injury models: identification of key genes involved in the development of kidney disease. Kidney Blood Press Res 2018;43:1898-907. [Crossref] [PubMed]
- Ke H, Zhang X, Cheng L, et al. Bioinformatic analysis to explore key genes associated with brain ischemia-reperfusion injury in rats. Int J Neurosci 2019;129:945-54. [Crossref] [PubMed]
- Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010;26:2363-7. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Janky R, Verfaillie A, Imrichova H, et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol 2014;10:e1003731. [Crossref] [PubMed]
- Fan Y, Siklenka K, Arora SK, et al. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res 2016;44:W135-41. [Crossref] [PubMed]
- Dehaini H, Awada H, El-Yazbi A, et al. MicroRNAs as potential pharmaco-targets in ischemia-reperfusion injury compounded by diabetes. Cells 2019. [Crossref] [PubMed]
- Liu S, He Y, Shi J, et al. Downregulation of miRNA-30a enhanced autophagy in osthole-alleviated myocardium ischemia/reperfusion injury. J Cell Physiol 2019. [Epub ahead of print]. [PubMed]
- Benstoem C, Stoppe C, Liakopoulos OJ, et al. Remote ischaemic preconditioning for coronary artery bypass grafting (with or without valve surgery). Cochrane Database Syst Rev 2017;5:CD011719. [PubMed]
- Bøtker HE, Lassen TR, Jespersen NR. Clinical translation of myocardial conditioning. Am J Physiol Heart Circ Physiol 2018;314:H1225-52. [Crossref] [PubMed]
- Hernandez-Resendiz S, Chinda K, Ong SB, et al. The role of redox dysregulation in the inflammatory response to acute myocardial ischaemia-reperfusion injury - adding fuel to the fire. Curr Med Chem 2018;25:1275-93. [Crossref] [PubMed]
- Opfermann P, Derhaschnig U, Felli A, et al. A pilot study on reparixin, a CXCR1/2 antagonist, to assess safety and efficacy in attenuating ischaemia-reperfusion injury and inflammation after on-pump coronary artery bypass graft surgery. Clin Exp Immunol 2015;180:131-42. [Crossref] [PubMed]
- Laffey JG, Boylan JF, Cheng DC. The systemic inflammatory response to cardiac surgery: implications for the anesthesiologist. Anesthesiology 2002;97:215-52. [Crossref] [PubMed]
- Hausenloy DJ, Erik Bøtker H, Condorelli G, et al. Translating cardioprotection for patient benefit: position paper from the Working Group of Cellular Biology of the Heart of the European Society of Cardiology. Cardiovasc Res 2013;98:7-27. [Crossref] [PubMed]
- Liakopoulos OJ, Schmitto JD, Kazmaier S, et al. Cardiopulmonary and systemic effects of methylprednisolone in patients undergoing cardiac surgery. Ann Thorac Surg 2007;84:110-8; discussion 118-9. [Crossref] [PubMed]
- Jun JH, Shim JK, Oh JE, et al. Protective effect of ethyl pyruvate against myocardial ischemia reperfusion injury through regulations of ROS-related NLRP3 inflammasome activation. Oxid Med Cell Longev 2019;2019:4264580. [Crossref] [PubMed]
- Gray CB, Suetomi T, Xiang S, et al. CaMKIIδ subtypes differentially regulate infarct formation following ex vivo myocardial ischemia/reperfusion through NF-κB and TNF-α. J Mol Cell Cardiol 2017;103:48-55. [Crossref] [PubMed]
- Sehgal PB, Helfgott DC, Santhanam U, et al. Regulation of the acute phase and immune responses in viral disease. Enhanced expression of the beta 2-interferon/hepatocyte-stimulating factor/interleukin 6 gene in virus-infected human fibroblasts. J Exp Med 1988;167:1951-6. [Crossref] [PubMed]
- Frei K, Malipiero UV, Leist TP, et al. On the cellular source and function of interleukin 6 produced in the central nervous system in viral diseases. Eur J Immunol 1989;19:689-94. [Crossref] [PubMed]
- Nakajima K, Martinez-Maza O, Hirano T, et al. Induction of IL-6 (B cell stimulatory factor-2/IFN-beta 2) production by HIV. J Immunol 1989;142:531-6. [PubMed]
- Podgoreanu MV, White WD, Morris RW, et al. Inflammatory gene polymorphisms and risk of postoperative myocardial infarction after cardiac surgery. Circulation 2006;114:I275-81. [Crossref] [PubMed]
- Burzotta F, Iacoviello L, Di Castelnuovo A, et al. Relation of the -174 G/C polymorphism of interleukin-6 to interleukin-6 plasma levels and to length of hospitalization after surgical coronary revascularization. Am J Cardiol 2001;88:1125-8. [Crossref] [PubMed]
- Anderson DR, Poterucha JT, Mikuls TR, et al. IL-6 and its receptors in coronary artery disease and acute myocardial infarction. Cytokine 2013;62:395-400. [Crossref] [PubMed]
- De Gennaro L, Brunetti ND, Montrone D, et al. Subacute inflammatory activation in subjects with acute coronary syndrome and left ventricular dysfunction. Inflammation 2012;35:363-70. [Crossref] [PubMed]
- Kobara M, Noda K, Kitamura M, et al. Antibody against interleukin-6 receptor attenuates left ventricular remodelling after myocardial infarction in mice. Cardiovasc Res 2010;87:424-30. [Crossref] [PubMed]
- Hartman MH, Vreeswijk-Baudoin I, Groot HE, et al. Inhibition of interleukin-6 receptor in a murine model of myocardial ischemia-reperfusion. PLoS One 2016;11:e0167195. [Crossref] [PubMed]
- Allegretti M, Bertini R, Cesta MC, et al. 2-arylpropionic CXC chemokine receptor 1 (CXCR1) ligands as novel noncompetitive CXCL8 inhibitors. J Med Chem 2005;48:4312-31. [Crossref] [PubMed]
- Frangogiannis NG, Entman ML. Chemokines in myocardial ischemia. Trends Cardiovasc Med 2005;15:163-9. [Crossref] [PubMed]
- Ulbrich F, Lerach T, Biermann J, et al. Argon mediates protection by interleukin-8 suppression via a TLR2/TLR4/STAT3/NF-κB pathway in a model of apoptosis in neuroblastoma cells in vitro and following ischemia-reperfusion injury in rat retina in vivo. J Neurochem 2016;138:859-73. [Crossref] [PubMed]
- Lefer AM, Johnson G 3rd, Ma XL, et al. Cardioprotective and endothelial protective effects of [Ala-IL8]77 in a rabbit model of myocardial ischaemia and reperfusion. Br J Pharmacol 1991;103:1153-9. [Crossref] [PubMed]
- Mezzaroma E, Toldo S, Farkas D, et al. The inflammasome promotes adverse cardiac remodeling following acute myocardial infarction in the mouse. Proc Natl Acad Sci U S A 2011;108:19725-30. [Crossref] [PubMed]
- Toldo S, Mezzaroma E, O'Brien L, et al. Interleukin-18 mediates interleukin-1-induced cardiac dysfunction. Am J Physiol Heart Circ Physiol 2014;306:H1025-31. [Crossref] [PubMed]
- Mauro AG, Mezzaroma E, Torrado J, et al. Reduction of myocardial ischemia-reperfusion injury by inhibiting interleukin-1 alpha. J Cardiovasc Pharmacol 2017;69:156-60. [Crossref] [PubMed]
- Prober DA, Edgar BA. Growth regulation by oncogenes--new insights from model organisms. Curr Opin Genet Dev 2001;11:19-26. [Crossref] [PubMed]
- Huang CY, Fujimura M, Noshita N, et al. SOD1 down-regulates NF-kappaB and c-Myc expression in mice after transient focal cerebral ischemia. J Cereb Blood Flow Metab 2001;21:163-73. [Crossref] [PubMed]
- Valiente-Alandi I, Potter SJ, Salvador AM, et al. Inhibiting fibronectin attenuates fibrosis and improves cardiac function in a model of heart failure. Circulation 2018;138:1236-52. [Crossref] [PubMed]
- Pang L, Cai Y, Tang EH, et al. Cox-2 inhibition protects against hypoxia/reoxygenation-induced cardiomyocyte apoptosis via Akt-dependent enhancement of iNOS expression. Oxid Med Cell Longev 2016;2016:3453059. [Crossref] [PubMed]
- Saito T, Rodger IW, Shennib H, et al. Cyclooxygenase-2 (COX-2) in acute myocardial infarction: cellular expression and use of selective COX-2 inhibitor. Can J Physiol Pharmacol 2003;81:114-9. [Crossref] [PubMed]
- Kwong W, Liuni A, Zhou K, et al. Cyclooxygenase inhibition and rosuvastatin-induced vascular protection in the setting of ischemia-reperfusion: a human in vivo study. Vascul Pharmacol 2015;71:159-65. [Crossref] [PubMed]
- Nijmeijer R, Krijnen PA, Assink J, et al. C-reactive protein and complement depositions in human infarcted myocardium are more extensive in patients with reinfarction or upon treatment with reperfusion. Eur J Clin Invest 2004;34:803-10. [Crossref] [PubMed]
- Sharif-Paghaleh E, Yap ML, Puhl SL, et al. Non-invasive whole-body detection of complement activation using radionuclide imaging in a mouse model of myocardial ischaemia-reperfusion injury. Sci Rep 2017;7:16090. [Crossref] [PubMed]
- Farrar CA, Zhou W, Lin T, et al. Local extravascular pool of C3 is a determinant of postischemic acute renal failure. FASEB J 2006;20:217-26. [Crossref] [PubMed]
- Brown KM, Kondeatis E, Vaughan RW, et al. Influence of donor C3 allotype on late renal-transplantation outcome. N Engl J Med 2006;354:2014-23. [Crossref] [PubMed]
- Zhou W, Patel H, Li K, et al. Macrophages from C3-deficient mice have impaired potency to stimulate alloreactive T cells. Blood 2006;107:2461-9. [Crossref] [PubMed]
- Lin Z, Lin H, Li W, et al. Complement component C3 promotes cerebral ischemia/reperfusion injury mediated by TLR2/NFκB activation in diabetic mice. Neurochem Res 2018;43:1599-607. [Crossref] [PubMed]
- Sharif-Paghaleh E, Yap ML, Meader LL, et al. Noninvasive imaging of activated complement in ischemia-reperfusion injury post-cardiac transplant. Am J Transplant 2015;15:2483-90. [Crossref] [PubMed]
- Duarte S, Hamada T, Kuriyama N, et al. TIMP-1 deficiency leads to lethal partial hepatic ischemia and reperfusion injury. Hepatology 2012;56:1074-85. [Crossref] [PubMed]
- Fang X, Tao D, Shen J, et al. Neuroprotective effects and dynamic expressions of MMP9 and TIMP1 associated with atorvastatin pretreatment in ischemia-reperfusion rats. Neurosci Lett 2015;603:60-5. [Crossref] [PubMed]
- Vogt L, Schmitz N, Kurrer MO, et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J Clin Invest 2006;116:2817-26. [Crossref] [PubMed]
- Huang X, Feng Z, Jiang Y, et al. VSIG4 mediates transcriptional inhibition of Nlrp3 and Il-1β in macrophages. Sci Adv 2019;5:eaau7426.
- Frank MM, Zuraw B, Banerji A, et al. Management of children with hereditary angioedema due to C1 inhibitor deficiency. Pediatrics 2016. [Crossref] [PubMed]
- Emmens RW, Baylan U, Juffermans LJ, et al. Endogenous C1-inhibitor production and expression in the heart after acute myocardial infarction. Cardiovasc Pathol 2016;25:33-9. [Crossref] [PubMed]
- Kristiansen M, Graversen JH, Jacobsen C, et al. Identification of the haemoglobin scavenger receptor. Nature 2001;409:198-201. [Crossref] [PubMed]
- Thomsen JH, Etzerodt A, Svendsen P, et al. The haptoglobin-CD163-heme oxygenase-1 pathway for hemoglobin scavenging. Oxid Med Cell Longev 2013;2013:523652. [Crossref] [PubMed]
- Liu X, Wei J, Peng DH, et al. Absence of heme oxygenase-1 exacerbates myocardial ischemia/reperfusion injury in diabetic mice. Diabetes 2005;54:778-84. [Crossref] [PubMed]
- Yang M, Kong DY, Chen JC. Inhibition of miR-148b ameliorates myocardial ischemia/reperfusion injury via regulation of Wnt/β-catenin signaling pathway. J Cell Physiol 2019;234:17757-66. [Crossref] [PubMed]
- Luo H, Wang J, Liu D, et al. The lncRNA H19/miR-675 axis regulates myocardial ischemic and reperfusion injury by targeting PPARα. Mol Immunol 2019;105:46-54. [Crossref] [PubMed]
- Xu X, Kriegel AJ, Jiao X, et al. miR-21 in ischemia/reperfusion injury: a double-edged sword? Physiol Genomics 2014;46:789-97. [Crossref] [PubMed]
- Wu MY, Yiang GT, Liao WT, et al. Current mechanistic concepts in ischemia and reperfusion injury. Cell Physiol Biochem 2018;46:1650-67. [Crossref] [PubMed]
- Zhang X, Sun R, Liu L. Potentially critical roles of TNPO1, RAP1B, ZDHHC17, and PPM1B in the progression of coronary atherosclerosis through microarray data analysis. J Cell Biochem 2019;120:4301-11. [Crossref] [PubMed]
- Qiu Z, Ye B, Yin L, et al. Downregulation of AC061961.2, LING01-AS1, and RP11-13E1.5 is associated with dilated cardiomyopathy progression. J Cell Physiol 2019;234:4460-71. [Crossref] [PubMed]
- Li L, Pan Z, Yang X. Key genes and co-expression network analysis in the livers of type 2 diabetes patients. J Diabetes Investig 2019;10:951-62. [Crossref] [PubMed]


