Analysis of lncRNA-mRNA expression pattern in male textile workers with noise-induced hearing loss
Introduction
Noise induced hearing loss (NIHL) is an occupational disease with high incidence worldwide, and more than 10% of workers in developed countries suffer from it. Statistics in the United States indicate that 25% of American factories have noise that could induce hearing loss, 49% to 70% of male miners aged 50–70 years have hearing impairment. Similarly, occupational noise hazard is also a major unsolved public health problem in China. There are approximately 10 million employees working in noise-excessive environments, of whom around 1 million suffer from occupational hearing loss of varying degrees.
Different individuals have different susceptibility to noise hazards. Even if exposed to the same level of noise, there will be significant differences in hearing impairment. In view of this, it is an urgent task to screen NIHL susceptible population and carry out early control measures in the current work of safety production and labor protection. Long non-coding RNAs (lncRNAs) are a group of RNAs with transcript greater than 200 nucleotides (nt) in length and have no recognizable coding recognizable coding proteins. They play an important role in gene regulation and thus affect cell homeostasis in different cellular processes, including cell proliferation, differentiation, apoptosis, and metastasis and cycle stability (1). Structurally, lncRNAs ranging in size from about 100 to 100,000 bases are spliced or uncut, polyadenylated or non-polyadenylated, nuclear or cytoplasmic, and usually transcribed by RNA polymerase II or III (2).
Recent reports have revealed that lncRNAs are involved in oxidative stress, including the possible influence on gene expression, and the expression of lncRNAs is related to the genetic susceptibility of NIHL (3,4). Simultaneously, lncRNAs exert vital roles in the development of forebrain auditory system, such as affecting neurogenesis, synaptogenesis, cell migration, cell type specification, neurite growth (5-7). Mounting evidence has demonstrated that lncRNAs have the potential to become a noninvasive biomarker of tumors (8-10), and may also be useful in the auditory system. LncRNAs can be detected in a variety of biological fluids, including urine and blood. These characteristics proved that lncRNAs can be used as an ideal noninvasive biomarker for the diagnosis and prognosis of hearing loss. This study was designed to identify the difference of serum lncRNAs expression between NIHL and exposure workers, and to speculate the target gene in view of noise. It would provide support for elucidating the molecular mechanism of noise affecting hearing loss.
Methods
Ethics statement
This study considered the use of secondary data analysis to encrypt private information about patients with NIHL. With regard to ethical requirements, the study is in line with the Helsinki Declaration and approved by the Ethics Committee of Jiangsu Center for Disease Control and Prevention.
Study design
This study collected biological samples and data of noise workers undergoing occupational health check-ups in Yizheng Chemical Fiber Company Limited (Yangzhou, China) from 2012 to 2014, and established noise workers database. Noise exposure means that occupational permissible exposure limit for 8 h weighted average are over 85 dB(A). The results of pure tone test were mostly bilateral symmetrical sensorineural hearing loss. Frequencies to be tested include speech frequencies (500, 1,000, 2,000 Hz) and high frequencies (3,000, 4,000, 6,000, 8,000 Hz). Hearing threshold of workers with noise induced hearing loss showed a “V” type subsidence at 3,000–6,000 Hz. The definition of normal hearing meant that the hearing threshold was not exceeded 25 dB at verbal frequencies and high frequencies.
In accordance with the purpose of the study, the subjects were divided into two groups: NIHL (noise-induced hearing loss cases) and Exposure (noise-exposed participants with normal hearing). The main criteria for screening NIHL cases were as follows: male; occupational noise exposure; sensorineural hearing loss; pure tone test with reduced hearing threshold; exclusion of other causes of nervous deafness. To increase the accuracy of screening results and exclude the effects of other diseases on the expression of lncRNA, subjects had no disease or health abnormalities except hearing loss in NIHL patients. The matching factors between NIHL cases and Exposure individuals included age, smoking, noise exposure intensity, noise exposure duration and the use of protective devices.
Extraction and purification of total RNA
In the screening stage of microarray analysis, total RNAs in blood were extracted using Norgen’s Total RNA Purification Kit (Norgen Biotech, Canada). Isolated RNAs quantification and purity were evaluated by NanoDrop ND-1000 spectrophotometer (Thermo Scientific, USA).
LncRNA microarray analysis
Total RNA samples were handed over to KangChen Bio-tech for detection of lncRNA and mRNA expression profiles on the platform of ArrayStar Human LncRNA V4.0. The obtained data were standardized using GeneSpring GX V12.1 (Agilent Technologies). The corrected P<0.05 and FC >5 were used to screen lncRNAs and mRNAs with differentially expressed. Background adjustment was needed before analysis. The background value was based on the point of the lowest signal intensity (non-specific sample-probe binding value) or the average absorbance value from the background signal of the whole chip’s non-hybridization point (11). The corrected data had negative values, which were removed by standard value or singular value discarding method and coefficient of variation method. The expression value of the gene closest to the gene to be filled was used to predict the expression value of the gene to be filled to fill the missing value. When processing data, in order to make the data in the same range, it can be normalized according to the mean and median (12).
Functional enrichment analysis
Gene ontology (GO) has annotated the functions of genes in detail and classified them into three categories: biological processes (BP), cellular components (CC), and molecular functions (MF). The differentially expressed genes found in the mRNA expression profiles were analyzed in the GO database (http://www.geneontology.org) (13). KEGG signal transduction pathway database (http://www.genome.jp/kegg/) were performed to clarify intuitively the role and significance of target genes in the overall biological pathway (14,15). Chi-square test was used to compare the proportion of genes related to a certain pathway in the target gene and the proportion of genes related to that pathway in the reference gene (P<0.05, FDR <0.05). The −log10 (P value) indicated the significance of GO enrichment in differentially expressed genes. FDR stood for the false discovery rate.
Quantitative real-time PCR
Total RNAs in plasma were extracted by TRIzol reagent (Invitrogen, USA). cDNAs were synthesized by reverse transcription using superscript III (Invitrogen, USA). Analysis of plasma lncRNA expression was analyzed by Quantitative real-time PCR with SYBR-Green. Quantitative PCR reactions were performed using ABI 7900 system with cycling requirement of 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. Each PCR board was set blank to exclude exogenous DNA and the contamination of cDNA during sample addition. The Quantitative PCR was carried out in triplicate. As illustrated in Table 1, GAPDH was used as the internal reference gene and specific primers were designed in the light of the full-length gene sequence.

Full table
Statistics
The research information was input into the EpiData database for summary analysis and quality check. Statistical analysis of the information was performed with SPSS 22.0 and GraphPad Prism 5.0. The matching of age, smoking, drinking, noise exposure duration and intensity was analyzed by student’s t-test and Pearson chi-square test. P values <0.05 represented statistical significance.
Results
General characteristics of the subjects
A total of 20 male workers were enrolled in the study, including 10 NIHL cases (mean age: 44.0±5.3) and 10 noise-exposed participants with normal hearing (mean age: 43.6±5.3). Statistical analysis displayed that there was no significant difference in age (P=0.868), smoking (P=0.582) and drinking (P=1.000) status as well as exposure years (P=0.844) between the two groups. No significant difference was obtained in the distribution of noise intensity among NIHL cases and controls (P=0.619), as shown in Table 2. The ultimate verification 6 lncRNAs characteristic information was displayed in Table 3.

Full table

Full table
Expression differences of lncRNA and mRNA via microarray analysis
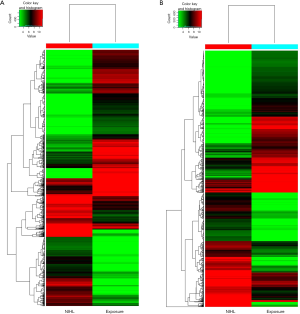
A total of 2,072 lncRNAs in two groups were significantly different (P<0.05, FC >5), 889 of which were up-regulated and 1,183 were down-regulated. As to mRNAs, 652 were up regulated and 811 were down regulated (P<0.05, FC >5). According to copies of lncRNA and amplification and expression of the primers, we selected six highly differentially expressed lncRNAs from 2,072 included lncRNAs, three of which were up-regulated (LOC101928211, LOC101929549, LOC101928804) and three were down-regulated (ZNF503-AS2, C1QTNF1-AS1, BANCR). Heat map, illustrated in Figure 1, were performed in the light of the expressed lncRNAs and mRNAs. Figure 1A elucidated that lncRNAs were recognized between NIHL and Exposure serum samples, while Figure 1B showed differently expressed mRNAs in two groups.
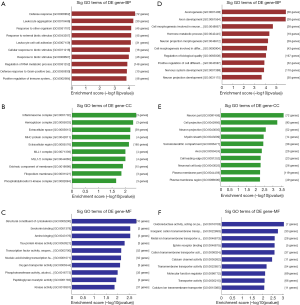
Functional analysis of differentially expressed mRNAs
In the GO analysis, by setting P<0.05, among the up-regulated mRNAs, the 10 most significant GO terms indicated there were 491 aberrantly expressed mRNAs allocated to biological process, 263 allocated to cellular component and 201 allocated to MF. The highest enrichment molecule metabolic process were defense response (GO: 0006952), inflammasome complex (GO: 0061702) and structural constituent of cytoskeleton (GO: 0005200) respectively. While 1425 differentially expressed mRNAs were down-regulated. The BP, CC and MFs with the highest enrichment were axonogenesis (GO: 0007409), neuron part (GO: 0097458) and oxidoreductase activity (GO: 0016709) separately, as shown in Figure 2.
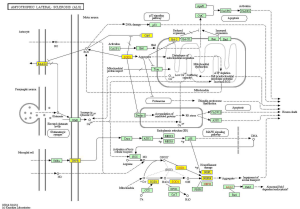
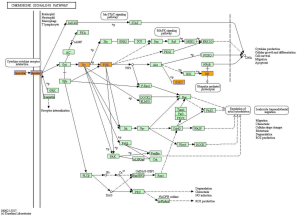
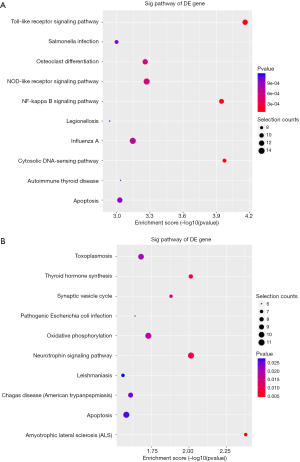
KEGG pathway analysis illuminated that there were 32 up-regulation pathways (P<0.05) and 18 down-regulation pathways (P<0.05). Figure 3 showed the top 10 up-regulation (Figure 3A) and down-regulation pathways (Figure 3B). 10 downward pathways contained 87 differential expression of genes, the most obvious one being Amyotrophic lateral sclerosis (ALS)-Homo sapiens (human) (hsa05014) (Figure 4). The up regulation of the 10 pathways contained 107 differential expression of genes, the most obvious of which was Toll-like receptor signaling pathway-Homo sapiens (human) (hsa04620) (Figure 5).
Identification of lncRNAs expression level by qRT-PCR
In this study, as chip detection used relatively limited number of sample, the stability of the detection results needs to be improved, and false positive results may be produced. To ensure a reliable differential expression of lncRNAs, 3 lncRNAs (LOC101928211, LOC101928804, and BANCR) were selected for qRT-PCR. Compared exposure group, the expression of LOC101928211 and LOC101928804 increased significantly in NIHL group, nevertheless, the expression of BANCR decreased significantly (P<0.001). The results were listed in Figure 6.

Discussion
NIHL is affected by both genetic and environmental factors. Our study focuses on the relationship between genetic factors and NIHL. Long-term exposure to noise can increase ROS and free radicals in the body's cochlea (16). In the case of excessive ROS or decreased antioxidant capacity of the body, free radicals can cause lipid peroxidation and damage to proteins, lipids, nucleic acids and other macromolecule substances, eventually leading to necrosis and apoptosis of cochlear cells. LncRNAs are not directly involved in protein expression, but it can regulate gene expression in the form of epigenetics, transcription and post-transcription levels. It has been confirmed that lncRNAs affect cell proliferation, apoptosis, and differentiation (17). By considering the mechanism of lncRNAs induced cell apoptosis and noise damage in cochlear hair cell, we look forward to serum lncRNAs as potential biomarker to indicate cochlear injury caused by excessive noise exposure, which provides a theoretical basis for early screening of NIHL susceptible population.
Hearing loss is related to heredity and environmental factors, including gender, age and noise exposure (18). In this study, 10 NIHL and 10 exposure subjects were enrolled. Merely male workers were included in the study owing to the serum samples of female noise exposed workers was less than 10% of all. Accordingly, female serum samples were not representative. In virtue of the influence of age on hearing function, all subjects were matched to exclude the influence of age on the study. At the same time, some studies (19-22) have illuminated that smoking or drinking increases the risk of NIHL. Consequently, smoking or drinking was included in the matching to reduce the impact of confounding factors.
LncRNAs have been found to carry many biological functions and play an important role in gene expression regulation. LncRNAs regulates gene expression in a variety of ways, mainly manifested in its dependence on gene expression regulation mechanisms. LncRNA plays a role not only as a major transcriptional regulator, but also as a co-regulator with other factors (6). A minority of lncRNAs have been explored to be associated with diseases, especially tumors. However, a key area in which lncRNAs has not been fully studied is the inner ear, which is responsible for hearing and homeostasis, and often leads to deafness and imbalance when it comes to disease. Most of the lncRNA is unreported, and so is in the study of NIHL. Wang et al. found that the differentially expressed lncRNA HOTAIR in serum might contribute to regulating the body’s response to noise-induced inner ear injury, in accordance with genotype specific role of rs4759314 in HOTAIR expression in hair cells (3). In addition, the expression of lncRNA HOTAIR can be used as an independent prognostic factor. HOTAIR can control cell cycle progression by interacting with EZH2. However, knocking out HOTAIR gene or EZH2 can inhibit cell cycle progression during cell culture (23). This suggests that when noise induces abnormal HOTAIR expression, it may affect the development cycle of cochlear hair cells and result in hearing loss. Ushakov et al. assessed the expression of selected lncRNAs in the two stages of hearing (E16.5 and P0). The results showed that more than 3,000 lncRNAs expressed in these stages of the inner ear had specific patterns of expression during the development of hair, sertoli cells and stria vascularis. In addition, a new group of lncRNAs related to deafness, including linc_Gata3, linc_miR96, was identified (4). GATA3, a zinc finger transcription factor, is believed to play a role in cochlear junction. Mutations in GATA3 are associated with deafness (24). The most common diseases include sensorineural hearing loss. MiR96 mutation causes progressive non-syndromic hearing loss in mammals (25). However, the underlying molecular mechanisms of this pathology have not been fully elucidated. Linc_miR96 contains pre-miRNA of miRNA 183/96/182 cluster in its intron. Both miRNA182 and miRNA183 are highly expressed during vestibule and cochlea development, Linc_miRNA96 thus may exert vital roles in the transcriptional regulation of miRNA96 loci. Exploring these differences in lncRNAs expression reinforces the hypothesis that these genomic transcripts play a key role as regulators of the auditory and vestibular systems.
To predict the potential function of lncRNAs, we used GO analysis and KEGG pathway annotation to study the co-expressed mRNAs. GO enrichment analysis revealed that the number of genes corresponding to regulation of up mRNAs were greater than that of down regulated mRNAs. In the light of GO analysis of differentially expressed genes, we discovered the enriched GO terms, and forecasted these genes may be related to the functional changes of different noise exposed populations (such as IL1B, HBA1, and INHA). The GO term, GO: 0006952, was related to the formation and development of the antioxidant defense system in organism tissues, affecting the activity of antioxidant enzymes and causing cell necrosis (26). Hearing loss was associated with necrosis of specific tissues, which further suggested that NIHL-related lncRNA may also be involved in cell survival and proliferation. KEGG pathway annotation indicated that there were 32 up regulation pathways and 18 down-regulation pathways. The significantly enriched up-regulated pathways like Toll-like receptor signaling pathway (hsa04620), Cytosolic DNA-sensing pathway (hsa04623), NF-kappa B signaling pathway (hsa04064) contained significantly up-regulated genes (CCL3L1, CCL4, AIM2, BCL2A1). Toll-like receptor (TLR) is activated in response to viral infection or stress (27). When TLR identifies pathogens, antigen presenting cells activate and stimulate signal transduction, leading to the production of various cytokines (28). Higher TLR2 expression may mean more severe cochlear injury in patients with sudden sensorineural hearing loss (SSNHL) (29). Therefore, functional analysis suggests that lncRNAs may play an important role in the pathogenesis of NIHL by regulating the corresponding signaling pathways. Overexpression of genes is conducive to cell proliferation, activation, differentiation, anti-apoptosis, immune response and transcription. The expression of lncRNAs with these genes might be contributed to regulation of these processes in chronic occupational hearing loss. However, this study has not explained the causal relationship between NIHL and serum lncRNAs expression. It indicated that they might be significant to the development of noise hearing loss related diseases.
By consulting relevant literature, we selected lncRNA with research background and potential functional significance for qRT-PCR. Compared with the noise-exposed group, the expression of two lncRNAs (LOC101928211, LOC101928804) was still significantly increased, and one lncRNA (BANCR) was significantly decreased in the NIHL group. QRT-PCR results confirmed our microarray data, indicating that the identified lncRNAs may play a crucial role in improving hearing loss. However, the study of lncRNAs in the inner ear is still in progress. There is no direct evidence that LOC101928211, LOC101928804, and BANCR are expressed in the inner ear. Therefore, we have searched for some possible evidence from studies on the pathogenesis of hearing loss and the functional expression of these three kinds of lncRNAs.
BANCR, as a tumor promoting gene, promotes proliferation and anti-apoptosis of gastric cancer cells, which was consistent with the expression in NIHL in this study. Zhang et al. detected the expression of BGC-823 in gastric cancer cells and GES-1 in normal gastric cancer epithelial cells, and found that BANCR was up-regulated in both gastric cancer tissues and cell lines (30). Down regulated BANCR regulates the proliferation and apoptosis of gastric cancer cell lines by inhibiting the expression of NF-κB1 (p50/105). It has been documented that BANCR could affect the proliferation of melanoma cells by regulating the activity of MAPK signaling pathway (ERK1/2) and (JNK) in vitro and in vivo (31) as well as implicated in cochlear hair cell cycle. All these studies demonstrated that differentially expressed lncRNAs may contribute to the regulation of noise-induced inner ear injury. There is no report on two lncRNAs (LOC101928211, LOC101928804) found in this study, our results preliminarily indicated that these two lncRNAs may be involved in the pathogenesis of NIHL.
Conclusions
In this preliminary study, we identified a group of expressed lncRNAs in the serum of patients with chronic occupational hearing loss. Together, our results suggested that screened lncRNAs might be involved in the pathological reaction and development of occupational noise related diseases, particularly in hearing loss. However, there are still many challenges posed by the heterogeneity of lncRNAs expression to be solved. Firstly, in this study, we intended to preliminarily understand the difference of serum lncRNA expression profiles between NIHL and control group, so the number of samples was small. Future studies will be conducted by recruiting more NIHL patients and exposed groups to confirm the mechanism of these multipurpose molecules and verify the clinical significance and diagnostic value of serum lncRNA, which will deepen our understanding of the pathogenesis of occupational hearing loss and provide new approaches for the diagnosis and treatment of occupational hearing loss.
Acknowledgments
Funding: Jiangsu Province’s Outstanding Medical Academic Leader program (CXTDA2017029).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of Jiangsu Center for Disease Control and Prevention.
References
- Zhang K, Shi H, Xi H, et al. Genome-Wide lncRNA Microarray Profiling Identifies Novel Circulating lncRNAs for Detection of Gastric Cancer. Theranostics 2017;7:213-27. [Crossref] [PubMed]
- Dinger ME, Amaral PP, Mercer TR, et al. Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Brief Funct Genomic Proteomic 2009;8:407-23. [Crossref] [PubMed]
- Wang B, Ding E, Shen H, et al. Association of TagSNP in lncRNA HOTAIR with susceptibility to noise-induced hearing loss in a Chinese population. Hear Res 2017;347:41-6. [Crossref] [PubMed]
- Ushakov K, Koffler-Brill T, Rom A, et al. Genome-wide identification and expression profiling of long non-coding RNAs in auditory and vestibular systems. Sci Rep 2017;7:8637. [Crossref] [PubMed]
- Guo Y, Zhang P, Sheng Q, et al. lncRNA expression in the auditory forebrain during postnatal development. Gene 2016;593:201-16. [Crossref] [PubMed]
- Aprea J, Prenninger S, Dori M, et al. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J 2013;32:3145-60. [Crossref] [PubMed]
- Berghoff EG, Clark MF, Chen S, et al. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development 2013;140:4407-16. [Crossref] [PubMed]
- Wang Y, Gu X, Li Z, et al. microRNA expression profiling in multidrug resistance of the 5Fuinduced SGC7901 human gastric cancer cell line. Mol Med Rep 2013;7:1506-10. [Crossref] [PubMed]
- Sánchez Y, Segura V, Marin-Bejar O, et al. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun 2014;5:5812. [Crossref] [PubMed]
- Xu C, Shao Y, Xia T, et al. lncRNA-AC130710 targeting by miR-129-5p is upregulated in gastric cancer and associates with poor prognosis. Tumour Biol 2014;35:9701-6. [Crossref] [PubMed]
- Brown CS, Goodwin PC, Sorger PK. Image metrics in the statistical analysis of DNA microarray data. Proc Natl Acad Sci U S A 2001;98:8944-9. [Crossref] [PubMed]
- Bolstad BM, Irizarry RA, Åstrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19:185-93. [Crossref] [PubMed]
- Malouf GG, Zhang J, Yuan Y, et al. Characterization of long non-coding RNA transcriptome in clear-cell renal cell carcinoma by next-generation deep sequencing. Mol Oncol 2015;9:32-43. [Crossref] [PubMed]
- Quénet D, Dalal Y. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife 2014;3:e03254. [Crossref] [PubMed]
- Tseng YY, Moriarity BS, Gong W, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014;512:82-6. [Crossref] [PubMed]
- Van Campen LE, Murphy WJ, Franks JR, et al. Oxidative DNA damage is associated with intense noise exposure in the rat. Hear Res 2002;164:29-38. [Crossref] [PubMed]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298-307. [Crossref] [PubMed]
- Lie A, Skogstad M, Johannessen HA, et al. Occupational noise exposure and hearing: a systematic review. Int Arch Occup Environ Health 2016;89:351-72. [Crossref] [PubMed]
- Palmer KT, Griffin MJ, Syddall HE, et al. Cigarette smoking, occupational exposure to noise, and self reported hearing difficulties. Occup Environ Med 2004;61:340-4. [Crossref] [PubMed]
- Shen H, Cao J, Hong Z, et al. A functional Ser326Cys polymorphism in hOGG1 is associated with noise-induced hearing loss in a Chinese population. PLoS One 2014;9:e89662. [Crossref] [PubMed]
- Tao L, Davis R, Heyer N, et al. Effect of cigarette smoking on noise-induced hearing loss in workers exposed to occupational noise in China. Noise Health 2013;15:67-72. [Crossref] [PubMed]
- Mohammadi S, Mazhari MM, Mehrparvar AH, et al. Cigarette smoking and occupational noise-induced hearing loss. Eur J Public Health 2010;20:452-5. [Crossref] [PubMed]
- Battistelli C, Cicchini C, Santangelo L, et al. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene 2017;36:942-55. [Crossref] [PubMed]
- Appler JM, Lu CC, Druckenbrod NR, et al. Gata3 is a critical regulator of cochlear wiring. J Neurosci 2013;33:3679-91. [Crossref] [PubMed]
- Mencía A, Modamio-Hoybjor S, Redshaw N, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet 2009;41:609-13. [Crossref] [PubMed]
- Ohtani H, Nakajima T, Akari H, et al. Molecular evolution of immunoglobulin superfamily genes in primates. Immunogenetics 2011;63:417-28. [Crossref] [PubMed]
- Marra CM, Sahi SK, Tantalo LC, et al. Toll-like receptor polymorphisms are associated with increased neurosyphilis risk. Sex Transm Dis 2014;41:440-6. [Crossref] [PubMed]
- Vethanayagam RR, Yang W, Dong Y, et al. Toll-like receptor 4 modulates the cochlear immune response to acoustic injury. Cell Death Dis 2016;7:e2245. [Crossref] [PubMed]
- Yang CH, Hwang CF, Yang MY, et al. Expression of toll-like receptor genes in leukocytes of patients with sudden sensorineural hearing loss. Laryngoscope 2015;125:E382-7. [Crossref] [PubMed]
- Zhang ZX, Liu ZQ, Jiang B, et al. BRAF activated non-coding RNA (BANCR) promoting gastric cancer cells proliferation via regulation of NF-kappaB1. Biochem Biophys Res Commun 2015;465:225-31. [Crossref] [PubMed]
- Li R, Zhang L, Jia L, et al. Long non-coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS One 2014;9:e100893. [Crossref] [PubMed]