
Comprehensive circular RNA profiling identifies CircFAM120A as a new biomarker of hypoxic lung adenocarcinoma
Introduction
Lung cancer is the most commonly diagnosed malignant tumor and the leading cause of cancer related death worldwide (1). In recent years, lung adenocarcinoma has become the dominant pathological type, especially in early staged lung cancer detected by computed tomography screening (2). The prognosis of lung cancer mainly depends on tumor metastatic status, and adenocarcinoma, in particular, may have a higher risk of metastasis than squamous cell carcinoma (3). On the other hand, the risk of metastasis also determines the treatment strategy for lung cancer, for instance, the extent of surgical resection and the choice of postoperative therapy (4,5). Therefore, assessing the risk of tumor metastasis is of great significance in promoting individualized treatment of lung cancer (6).
Hypoxia in cancer is a local and tumor-specific micro-environmental change caused by imbalance between increased oxygen demand from rapidly growing tumor cells and relatively insufficient blood supply (7). Tumor hypoxia is well recognized as a key promoting factor in the initiation and progression of metastasis (7-9), and it is common in lung cancer (10,11). Enatsu et al. found that the expression of Hypoxia-Inducible Factor-1α (HIF-1α) assessed by immunohistochemistry was significantly elevated in lung cancer tissue, which was correlated with lymph node metastasis and lympho-vascular invasion status (12). The 5-year survival rate of lung adenocarcinoma was significantly lower in patients with high HIF-1α expression (8,12). Therefore, biomarkers of lung cancer hypoxia may predict the risk of tumor metastasis. Unfortunately, there is currently no clinical indicator that can fully reflect the degree of tumor hypoxia.
Circular RNA (circRNA) is a group of endogenous non-coding RNA which are mainly enriched in the cytoplasm and exosomes (13,14). CircRNAs are mainly generated by ‘back-splicing’ processors and have neither 5'-3' polarity nor a polyadenylation tail, which renders them resistant to degradation by RNase R and therefore more stable than linear RNA (13). Recent studies have shown that circRNAs are involved in both physiological and pathological processes via multiple actions, for example binding to other ribonucleic acids such as mRNA and miRNA as molecular sponge (13,15,16). CircRNA profiles can also dynamically reflect phenotypical changes of the cell (12,15). Notably, due to its stability, tumor-specific circRNAs can be detected in various body fluids including blood and urine, and have the potential to become novel tumor biomarkers (17,18). At present, the impact of hypoxia on the expression of circRNAs in lung cancer is still unknown.
This study aims to characterize circRNA profiles of the hypoxic lung adenocarcinoma cells and identify new biomarkers for the assessment of tumor hypoxia, thus may inspire future development of novel indicator of lung adenocarcinoma metastatic risks.
Methods
Human samples and online data mining
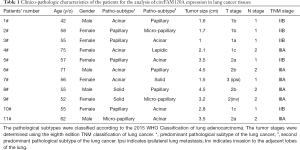
Paired tumor specimens and adjacent normal lung tissue were collected from patients with lung adenocarcinoma during the surgery and immediately stored in liquid nitrogen. All the patients received radical lobectomy with systemic lymph node dissection at Shanghai Chest Hospital between January 2018 and July 2018. None of these patients had preoperative chemotherapy or radiotherapy. Clinico-pathologic features was shown in Table 1. This study was approved by the Clinical Research Ethics Committee of Shanghai Chest Hospital. Written informed consent was obtained from all participants.
Full table
Clinical data for lung adenocarcinoma patients were downloaded from the Kaplan-Meier Plotter (http://kmplot.com/analysis/index.) and gene expression data are downloaded from TCGA database.
Cell cultures
A549 cell were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin and cultured at 37 °C, 21% O2 and 5% CO2 in a standard and humidified tissue culture chamber (normoxia). For hypoxia studies, the cells were cultured at 37 °C under 1% O2 and 5% CO2 in a humidified hypoxic chamber (Forma Scientific, Marietta, OH, USA).
CCK8 assay
A549 cells were cultured under normoxia (21% O2, 5% CO2) or hypoxia (1% O2, 5% CO2) for 4h according to previous reports (19,20) and returned to normoxia for 24 or 48 hours. 10 µL of CCK8 solution was added to A549 cells and incubated for 4 h. The absorbance at 450 nm was measured in a microplate reader.
Wound-healing assay
A549 cells (5×105) were seeded into 35 mm dish and cultured till 100% confluent. Prior to wounding, the cells were cultured under normoxia (21% O2, 5% CO2) or hypoxia (1% O2, 5% CO2) for 4h. Next, a sterile pipette tip (1 mL) was used to generate a wound, which was vertical to the parallel line (the interval was 2–3 mm) marked on the bottom of the dish and then washed with PBS for three times to remove cell debris. Images were taken at 0, 24 and 48 hours after the scratches.
Transwell assay
Transwell chamber (24-well, 8.0-µm pore membranes, Corning, USA) was used according to the manufacturer’s protocol. A549 cells (1×105 per well) were seeded in the upper chamber in 200 µL of serum-free medium, and 600 µL of complete medium was added to the lower chamber as a chemoattractant at the same time. After incubated under normoxia or hypoxia for 4 hours, the cells remaining on the upper surface of the membrane were removed. Migrated cells on the lower surface of the membrane were fixed with 4% paraformaldehyde, stained with hematoxylin and eosin, and photographed by inverted fluorescence microscope.
Western blot analysis
Proteins were extracted with RIPA buffer consisting 50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM PMSF, 10 mM DTT and protease inhibitors. The protein lysates were collected using a scraper and separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred into a NC membrane (PALL, 66485). After blocking with 5% skimmed milk, the membrane was incubated overnight at 4 °C with HIF-1α (Abcam, ab51608) or α-tubulin (Santa Biotechnology, sc-398103) primary antibodies. Subsequently, the membrane was incubated with secondary antibodies at room temperature, including Goat anti-Mouse (LI-COR, 926-68070) and Goat anti-Rabbit (LI-COR, 926-68071). The images of western blots were collected and analyzed by Odyssey CLx Imaging System (LI-COR).
RNA extraction and quality control (QC)
Total RNA of each sample was isolated using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol. The quantity and quality of total RNA samples were measured using NanoDrop ND-1000 (Thermo Scientific, USA). RNA integrity was tested by denaturing 1% gel electrophoresis. RNA samples were stored at −80 °C before use.
RNA library construction and circRNA sequencing
RiboRNAs in total RNA was removed using Ribo-Zero rRNA Removal Kits (Illumina, USA). RNA libraries were constructed using rRNA-depleted RNAs with TruSeq Stranded Total RNA Library Prep Kit (Illumina, USA) according to the manufacturer’s protocol. The RNA sequencing library was evaluated by the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 System (Agilent Technologies). Total RNA from four matched samples were treated with Epicenter Ribo-Zero rRNA Removal Kit (Illumina, CA, USA) and RNAse R (Epicenter, CA, USA) to remove ribosomal and linear RNA. Then, the RNA-seq libraries were constructed using TruSeq® Stranded Total RNA HT/LT Sample Prep Kit (Illumina, CA, USA). Sequencing was determined on Illumina Hiseq 2500 instrument. All the sequencing procedures and analyses were performed by RiboBio (Guangzhou, China).
Identification and quantification of human circRNAs
For each sample, FASTQ reads were first mapped to human reference genome (GRCh37/hg19) obtained from the UCSC genome database (http://genome.ucsc.edu/). The detected circRNAs annotated with the circBase database. Differentially expressed circRNAs were identified by Student’s-t tests and the fold change between the two groups. P-values were corrected by using multiple hypothesis testing (BH method). CircRNAs exhibiting fold changes ≥2.0 with P values ≤0.05 were classified as significant. The coordinates of the mRNA (which is defined as the circRNA-associated gene) were found through the RefSeq database based on back-splicing site coordinates of the circRNAs. Alignment was done by TopHat2 and differential expression analysis on RNA-seq data was achieved with the R package EBSeq. GO (http://www.geneontology.org) enrichment analysis and KEGG (http://www.genome.jp/kegg) pathway analysis was performed using the DAVID website (https://david.ncifcrf.gov/) for the differentially expressed circRNA-associated genes (21,22). A P value <0.05 was considered significant. After obtaining the predicted sequence of the circRNA, circRNA and miRNA interactions were predicted using customized miRNA target prediction software based on Miranda and RNAhybrid. The miRNA binding sites on differentially expressed circRNAs and the putative target miRNAs for differentially expressed circRNAs were identified. The circRNA-miRNA network was constructed using Cytoscape software 3.6.1.
Validation of circRNAs with polymerase chain reactions (PCRs), Quantitative real time-PCR (qRT-PCR), Sanger sequencing and Nuclear/cytosol isolation
To confirm the dysregulated circRNAs predicted by the sequencing analysis, 500 ng of total RNA was reverse-transcribed into cDNA with a random primer by using the SuperScript™ IV First-Strand Synthesis System (Invitrogen, USA) according to the manufacturer’s instructions. A set of divergent primers were designed on the flanking sequences of head-to-tail splicing sites of circRNAs. Polymerase chain reactions (PCRs) were done using these divergent primers and cDNA templates. Specific primers designed for the selected circRNAs were listed in Table S1. The human β-Actin reference gene was used to normalize the RNA levels. PCR products were separated using agarose gel electrophoresis, and purified with QIAquick Gel Extraction Kit (Qiagen, CA, USA). Sanger sequencing were performed to further confirm the presence of the back-spliced junction sites. For qRT-PCR, approximately 1 µg of RNA was used for cDNA synthesis using First-Strand cDNA Synthesis kit (Bio-Rad, Hercules, CA, USA). The qRT-PCR was performed using SYBR Green PCR Master Mix (Anzy Biotechnology, Shanghai, China). All measurements were performed in triplicate and standardized to the levels of β-actin. The primers of target genes were shown in Table S1. For the assessment of subcellular localization of circFAm120A, A549 nuclear and cytosol were separated with NE-PER Nuclear and Extraction Reagents (Thermo Scientific, CA, USA). QRT-PCR analysis was performed subsequently.

Full table
Statistical analysis
Statistical analyses were performed using SPSS 20.0 (IBM, SPSS, Chicago, IL, USA). Data were expressed as the mean ± SEM. Student’s t-test was used to determine the statistical significance for comparison of two groups. All tests were 2-sided, and P<0.05 was considered statistically significant.
Results
Phenotypic changes in A549 cells after hypoxic treatment
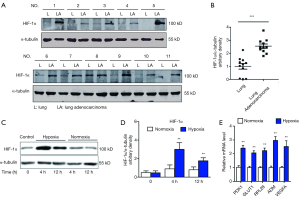
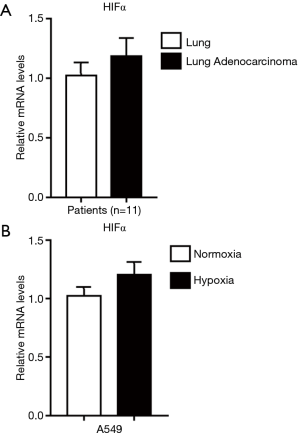
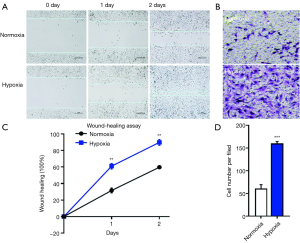
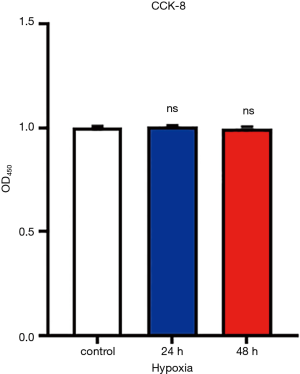
In lung adenocarcinoma tissues, HIF-1α expressions were significantly elevated as compared to the adjacent normal lung controls, suggesting the existence of tumor hypoxia during the development of lung adenocarcinoma, which was consistent with previous reports (8,10) (Figure 1A,B). Thus, we continued to study the contribution of hypoxia on circRNA profile alternations in A549 lung adenocarcinoma cell line. We firstly treated A549 lung adenocarcinoma cell line with normoxia or hypoxia. To check whether hypoxic phenotype could be successfully induced, the expression of HIF-1α and its downstream genes were examined in A549 cells after up to 12 hours of hypoxia (1% O2) treatment, the protein levels of HIF-1α (Figure 1C,D) and its target genes (18) (Figure 1E), such as pyruvate dehydrogenase kinase (PDK1), glucose transporter 1 (GLUT1), adrenomedullin (ADM), vascular endothelial growth factor A (VEGFA) and ribosomal protein L28 (RPL28) were significantly elevated as compared to independent A549 cultured under normoxia. Interestingly, HIF-1α mRNA levels were not changed, suggesting the hypoxia regulation on HIF-1α was on translational level (Figure S1). Consistently, cell migration was also stimulated by hypoxia treatment in A549 cells (Figure 2). After cultured under hypoxia, transverse and vertical migration rates of the lung cancer cells were both enhanced as confirmed by wound healing assay and Transwell migration assay respectively. Meanwhile, hypoxia treatment did not increase their proliferation as shown by CCK8 analysis (Figure S2), suggesting a pro-metastatic phenotype was induced in the treated lung adenocarcinoma cells (19).
Expression pattern of circRNAs in A549 lung adenocarcinoma cell lines
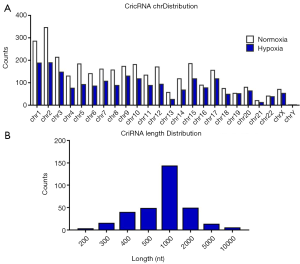
Next, high-throughput sequencing was used to detect the expression profile of circRNAs. We first characterized circular RNA transcripts using RNA-seq analysis of ribosomal RNA-depleted total RNA from four paired A549 cells. Each sample was sequenced on an Illumina HiSeq and the yielded reads were mapped to the human reference genome (GRCh37/hg19) by TopHat2. A computational pipeline based on the anchor alignment of unmapped reads was used to identify circRNAs without reliance on gene annotations. The algorithm utilizes unmapped reads as templates and aligns 20 nucleotides from either end of the reads, called “anchor sequences”, which were further extended. The reads with flanking splice site GU/AG were included as potential circular RNA reads (23-25). The Chr distribution of circRNA candidates which were found in two groups was shown in Figure 3A. Overall, a total of 558 circRNAs were identified in both groups. The size of circRNA candidates ranged from under 100 nt to over 2,000 nt. Approximately 75% of circRNAs had the predicted spliced length of less than 2,000 nt, including 45.56% of circRNAs less than 500 nt and 21.43% of circRNAs between 500 nt and 1,000 nt in length (Figure 3B).
Differential expression of circRNAs in A549 cells under normoxia and hypoxia conditions
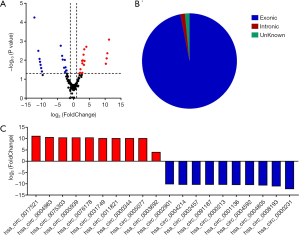
CircRNAs were sequenced in A549 cells cultured under normoxia and hypoxia conditions. The Volcano plot filtered and identified the differentially expressed circRNAs with statistical significance between the two groups (Figure 4A). The threshold of exhibiting folds change is 2.0 and P values <0.05. Among 65 identified circRNAs that were differentially expressed, 35 were up-regulated and 30 were down-regulated (Table S2). In light of the relationship between circRNAs and their coding genes, the circRNAs were summarized and classified into two categories: 63 were exonic, 2 were intronic (Figure 4B). According to the log fold change, the top ten upregulated and downregulated circRNAs were selected (Figure 4C).
Full table
Construction of a circRNA-miRNA network
Previous reports have shown that circRNAs could function as miRNA sponges to regulate the expression levels of other related RNAs by miRNA response elements (MRE) (11). Therefore, it is important to identify the interaction between circRNAs and miRNAs. Potential miRNA with MRE containing the binding sites that may be sponged by differentially expressed circRNAs were predicted to construct a circRNA-miRNA network (Figure 5A). According to this network, the top six predicted miRNA targets of each differentially expressed circRNAs were hsa-miR-12119, hsa-miR-370-3p, hsa-miR-370-3p, hsa-miR-4539, hsa-miR-6511a-5p, hsa-miR-6772-3p and hsa-miR-9851-3p. We then selected these miRNAs and 23 circRNAs, which had greater interaction with each other, to construct another more focused circRNA-miRNA network (Figure 5B). This network showed that seven circRNAs, including hsa_circ_0017521, hsa_circ_0003692, hsa_circ_0000615, hsa_circ_0001868, hsa_circ_0005939, hsa_circ_0008193 and hsa_circ_0001550, were changed under hypoxia with higher significance compared to normoxia conditions (P<0.01).
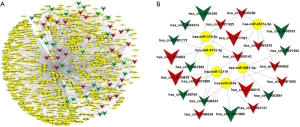
Identification of circFAM120A as potential biomarker of lung cancer hypoxia by qRT-PCR
Subsequently, hsa_circ_0008193, which was downregulated in the hypoxic A549 cells (Figure 4A) and also more miRNA interactive (Figure 5B), was chosen for further evaluation. CircFAM120A is located at chromosome 1q21 and is composed of three exons. We performed PCR analysis with divergent and convergent primers on circFAM120A by agarose gel detection after RNase H treatment (Figure 6A). The results showed that the PCR product was formed only with divergent primers, suggesting that circFAM120A was a circular RNA rather than a liner RNA (Figure 6B). Meanwhile, sanger sequencing also revealed the splicing site of circFAM120A (Figure 6C). Interestingly, circFAM120A predominantly exists in the cytosol but not in the nucleus (Figure 6D). Importantly, as validated by qRT-PCR in an independent set of A549 cells, the expression levels of CircFAM120A was significantly decreased after treatment with hypoxia (Figure 6E), which confirmed the high-throughput sequencing results.
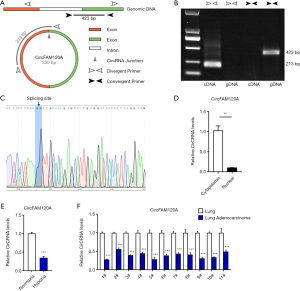
It is known that hypoxia is common in lung cancer with lymph node metastasis (Figure 1) (8,10). In order to explore whether CircFAM120A could be a good candidate biomarker of hypoxic lung cancer, we then examined its levels in eleven paired tumor tissues and adjacent non-cancer tissues from patients with lymph node positive lung adenocarcinoma (Table 1) using qRT-PCR. Consistent with the results from cell experiments, circFAM120A expression were significantly lower in all the cancer specimens (Figure 6F), implicating its potential value in distinguishing hypoxic lung adenocarcinoma. Interestingly, the expression level of the host gene FAM120A, although reported to be elevated in colon cancer (26), was not altered in lung adenocarcinoma patients from the TCGA database, which was also confirmed in our cohort (Figure S3).

GO enrichment and KEGG pathway analysis of CircFAM120A’s target mRNAs
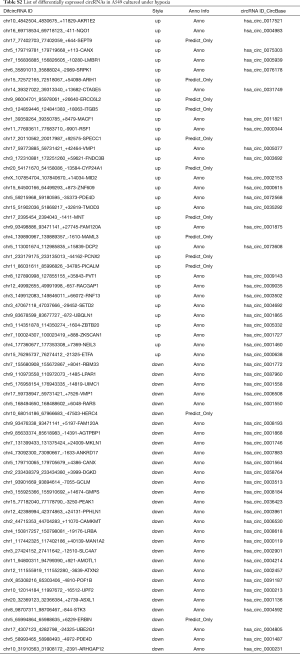
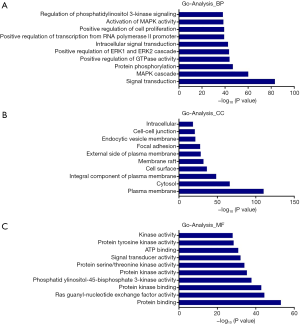
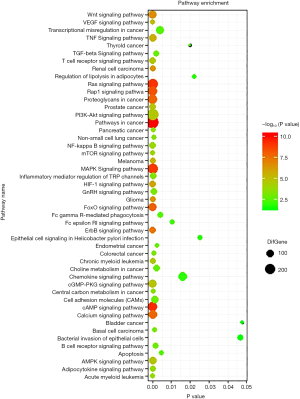
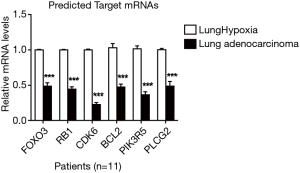
The target miRNAs of CircFAM120A (Table 2) and their downstream mRNA were predicted by Miranda and RNAhybrid (http://fp.amegroups.cn/cms/atm.2019.08.79-1.pdf). These target genes were used for GO enrichment analysis to annotate and speculate the function of these target genes. GO enrichment analysis is divided into three parts: biological process (BP), cell component (CC) and molecular function (MF). GO enrichment analysis of BP (Figure 7A) showed that these target genes were significantly associated with signal transduction and protein phosphorylation pathways such as MAPK cascade and ERK1/2 cascade. GO enrichment analysis of CC (Figure 7B) showed that these genes were mainly components of plasma membrane and cell junction proteins. As for molecular functions (Figure 7C), the regulation of protein binding, Ras guanyl-nucleotide exchange factor activity and protein kinase binding were the top predicted cell functions that may be affected by the altered circFAM120A expression. Consistently, the KEGG pathway dot plot showed the significantly enriched pathways with the top enrichment score (Figure 8). Among them, Pathways in cancer had the higher enrichment score as compared to the other signaling pathways. These results implicate that circFAM120A may have multiple biological functions and its dysregulation is likely to participate in the development of lung adenocarcinoma. Indeed, we examined and found decreased levels of a few predicted mRNA targets in lung adenocarcinoma specimen, including FOXO3, RB1, CDK6, BCL2, PIK3R5 and PLCG2 (Figure S4), which were consistent with the bioinformatic prediction (http://fp.amegroups.cn/cms/atm.2019.08.79-1.pdf).

Full table
Discussion
In this study, we investigated the effects of hypoxia on lung adenocarcinoma cells by circRNA high-throughput sequencing and further identified a circRNA marker (CircFAM120A) of tumor hypoxia which may have the potential to predict metastatic risk of lung cancer. The downstream functions and pathways of CircFAM120A were predicted by computational analysis. These results may shed a light on the development of novel lung cancer treatment strategy based on tumor hypoxic status.
Recently, the concept of individualized cancer therapy, which is to select treatment according to distinct characteristics of the tumor to reduce side effects, has gained increasing acceptance (5,6). Previously, we found intraoperative pathological subtyping may help surgeons select patients with pre-invasive lung cancer for limited lung and lymph node resections (4,5). However, there are still two main problems in this method: First, intraoperative pathological accuracy is not ideal and significantly prolongs the operation time. Second, most patients still have different prognosis even if their tumor size and pathology are identical, suggesting other factors may also affect tumor malignancy (4,27). Therefore, it is necessary to search for new biomarkers which can further stratify the patients before the surgery.
Hypoxia is a key micro-environmental factor that promotes tumor metastasis (7). Consistent with previous studies (9,10,28), we showed that in A549 cells cultured under 1% oxygen the expression of HIF-1a and its downstream genes were significantly elevated, leading to enhanced tumor migration ability. Hypoxia is very common in lung cancer (10,11). Even in early-stage surgically respectable lung cancer, the oxygen partial pressure of the lung cancer tissue is only about 15% of the surrounding lung tissue (8). In human and animal tumor specimens, cell invasion and epithelial-mesenchymal transition (EMT)-related genes were significantly enhanced in the hypoxic regions (9). The levels of tumor hypoxia-associated proteins in surgical specimen are significantly correlated with postoperative metastasis and recurrence rate (8-10). However, there is currently no clinical means to accurately assess the degree of tumor hypoxia. Although HIF-1α is often used as an indicator of tumor hypoxic status (7), its measurement mainly depends on the immunohistochemical evaluation of tumor histopathology which can only be obtained after the surgery, thus has no use in preoperative decision-making. Hypoxia tracing positron emission tomography can be used to assess the degree of tumor hypoxia in vivo, but this method is poorly stable, difficult to quantify, expensive and cause radioactive damage to the patients thus is hard to be promoted in clinic (29). Therefore, molecular markers that can be used for liquid biopsy and can reliably, comprehensively and dynamically reflect the degree of tumor hypoxia are ideal indicators for individualized treatment (29).
CircRNA, a group of non-coding RNAs, is characterized by covalently closed continuous loop without 5' to 3' polarity and polyadenylated tail. Because of its structural specificity, circRNA is resistant to RNase R digestion and more stable than other linear RNAs such as mRNA and miRNA (11). The half-life of circRNA is over 48 hours (11,13). Moreover, circRNAs can be secreted to extracellular space via exocytosis and is detectable in body fluids such as blood and urine (12,15,30). These properties render circRNAs with the potential to be ideal biomarkers for diagnosis of disease, including cancers (12,15). Zhang et al. demonstrated that ciRS-7 is elevated in lung cancer tissues and that the degree of elevation is associated with lymph node metastasis and poor prognosis (31). Tan et al. found that the circular RNA F-circEA-4a produced by the EML4-ALK fusion mutation can be secreted from the tumor to the peripheral blood and stably detected (32). In addition, Li et al. found that circRNAs are more enriched in exosomes and plays biological role in recipient cells. The levels of blood tumor circRNAs correlates with the prognosis of colorectal cancer (15). All these evidences suggest the potential of circRNA as a biomarker for tumor malignancy.
To our knowledge, studies investigating the impact of hypoxia on circRNA expressions in cancer cells are still in scarce. In breast cancer cells, hypoxia can up-regulate the expression of circDENND4c through HIF-1α and promote tumor cell proliferation (33). However, no such results have been reported in other cancers by far, including lung cancer. The high-throughput sequencing technology largely accelerates the discovery of the new molecules and drugs. Using microarray assay, Boeckel found that hypoxia cause significant changes in global circRNAs expression in human umbilical endothelial cells, and they further identified circZNF292, which was upregulated under hypoxia, was associated with vascular tube formation and spheroid sprouting of endothelial cells (34). In the present study, using a similar approach, we found a total of 65 circRNAs that are differentially expressed in lung adenocarcinoma under hypoxia, including 35 up-regulated and 30 down-regulated circRNAs. Bioinformatic analysis indicated some of these circRNAs may function as molecular sponge to miRNAs and affect the expression their downstream gene expression, suggesting their roles in the development of cancer under hypoxia. It has to be noted that we utilized bioinformatic tools TopHat2 and R package EBSeq to process the sequencing data while other bioinformatic tool such as HISAT2 may be superior in some aspects.
In the current study, we also identified that CircFAM120A, which was centered in the circRNA-miRNA network, was significantly downregulated in both lung adenocarcinoma cell line and tissues from the patients with lymph node metastasis by qRT-PCR. GO enrichment and KEGG pathways analyses predicted that CircFAM120a may affect cell membrane protein binding and signal transduction via the regulation of the targeted miRNA and mRNA, and is very likely to be involved in cancer development. Indeed, we found decreased levels of several known tumor suppressor genes such as FOXO3, RB1, CDK6, BCL2, PIK3R5 and PLCG2 (35), in the lung adenocarcinoma specimen compared to adjacent lung tissue. Future work on the validation of miRNA of circFAM120A would be more informative. Meanwhile, more detailed mechanisms of circFAM120A downregulation in lung adenocarcinoma specimen and its potential binding with putative miRNAs with Chromatin Isolation by RNA Purification (ChRIP) analysis still need future investigations. CircFAM120A is located on chromosome 9q22.31, which is aligned in a sense orientation to a known protein-coding gene FAM120A. FAM120A encodes intracellular cytoskeletal proteins and activates the downstream Src pathway under oxidative stress, therefore it is also known as oxidative stress-associated Src activator (Ossa) (21,36). Interestingly, activation of FAM120A promote proliferation and metastasis in colon cancer (21). However, we found that FAM120A mRNA levels were not altered in lung adenocarcinoma specimen in TCGA database and in our cohort, suggesting the potential specific roles of FAM120A in different cancer types and independence of expression levels of circFAM120A and its host gene FAM120 in lung adenocarcinoma. As circular RNA biogenesis may compete with pre-mRNA splicing (37), whether the downregulation of CircFAM120A is a consequence of increased oxidative stress and FAM120A activation under hypoxia merits further investigation.
In summary, the present study explored changes in circRNA profiles of lung cancer cells under hypoxia and identified circFAM12A as a potential biomarker of tumor hypoxia. It is hoped these results may provide new insights to the mechanism of hypoxia induced lung cancer metastasis, and promote individualized treatment that guided by the degree of tumor hypoxia.
Acknowledgments
The authors would like to thank Prof. Xinran Ma from the Institute of Biomedical Sciences and School of Life Sciences, East China Normal University, and his lab members for providing precious advices and technical supports to this project.
Funding: This research was supported by grants from the National Natural Science Foundation of China (81502480 to X Cheng and 31800989 to L Xu), Shanghai Hospital Development Center Grant (SHDC12016113 to Q Luo), the China Postdoctoral Science Foundation (2017M611499 to D Wang), Science and Technology Commission of Shanghai Municipality (18441901500 to X Cheng), Shanghai Municipal Health Commission (2018YQ61 to X Cheng), Shanghai Pujiang Program (17PJ1402700 to L Xu) and ECNU public platform for Innovation (011).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The authors have no conflicts of interest to declare. This study was approved by the Clinical Research Ethics Committee of Shanghai Chest Hospital. Written informed consent was obtained from all participants.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Yang W, Qian F, Teng J, et al. Community-based lung cancer screening with low-dose CT in China: Results of the baseline screening. Lung Cancer 2018;117:20-6. [Crossref] [PubMed]
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- Cheng X, Zheng D, Li Y, et al. Tumor histology predicts mediastinal nodal status and may be used to guide limited lymphadenectomy in patients with clinical stage I non-small cell lung cancer. J Thorac Cardiovasc Surg 2018;155:2648-56.e2. [Crossref] [PubMed]
- Liu S, Wang R, Zhang Y, et al. Precise Diagnosis of Intraoperative Frozen Section Is an Effective Method to Guide Resection Strategy for Peripheral Small-Sized Lung Adenocarcinoma. J Clin Oncol 2016;34:307-13. [Crossref] [PubMed]
- Cheng X, Onaitis MW, D'Amico T A, et al. Minimally Invasive Thoracic Surgery 3.0: Lessons Learned From the History of Lung Cancer Surgery. Ann Surg 2018;267:37-8. [Crossref] [PubMed]
- Rankin EB, Giaccia AJ. Hypoxic control of metastasis. Science 2016;352:175-80. [Crossref] [PubMed]
- Le QT, Chen E, Salim A, et al. An evaluation of tumor oxygenation and gene expression in patients with early stage non-small cell lung cancers. Clin Cancer Res 2006;12:1507-14. [Crossref] [PubMed]
- Popper HH. Progression and metastasis of lung cancer. Cancer Metastasis Rev 2016;35:75-91. [Crossref] [PubMed]
- Liu KH, Tsai YT, Chin SY, et al. Hypoxia Stimulates the Epithelial-to-Mesenchymal Transition in Lung Cancer Cells Through Accumulation of Nuclear β-Catenin. Anticancer Res 2018;38:6299-308. [Crossref] [PubMed]
- Nurwidya F, Takahashi F, Kobayashi I, et al. Treatment with insulin-like growth factor 1 receptor inhibitor reverses hypoxia-induced epithelial-mesenchymal transition in non-small cell lung cancer. Biochem Biophys Res Commun 2014;455:332-8. [Crossref] [PubMed]
- Enatsu S, Iwasaki A, Shirakusa T, et al. Expression of hypoxia-inducible factor-1 alpha and its prognostic significance in small-sized adenocarcinomas of the lung. Eur J Cardiothorac Surg 2006;29:891-5. [Crossref] [PubMed]
- Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet 2016;32:309-16. [Crossref] [PubMed]
- Vo JN, Cieslik M, Zhang Y, et al. The Landscape of Circular RNA in Cancer. Cell 2019;176:869-81.e13. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:981-4. [Crossref] [PubMed]
- Beermann J, Piccoli MT, Viereck J, et al. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev 2016;96:1297-325. [Crossref] [PubMed]
- Phillips RJ, Mestas J, Gharaee-Kermani M, et al. Epidermal growth factor and hypoxia-induced expression of CXC chemokine receptor 4 on non-small cell lung cancer cells is regulated by the phosphatidylinositol 3-kinase/PTEN/AKT/mammalian target of rapamycin signaling pathway and activation of hypoxia inducible factor-1alpha. J Biol Chem 2005;280:22473-81. [Crossref] [PubMed]
- Orlowski K, Rohrer Bley C, Zimmermann M, et al. Dynamics of tumor hypoxia in response to patupilone and ionizing radiation. PLoS One 2012;7:e51476. [Crossref] [PubMed]
- Barben M, Ail D, Storti F, et al. Hif1a inactivation rescues photoreceptor degeneration induced by a chronic hypoxia-like stress. Cell Death Differ 2018;25:2071-85. [Crossref] [PubMed]
- Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer 2014;14:430-9. [Crossref] [PubMed]
- Sharma D, Sehgal P, Mathew S, et al. A genome-wide map of circular RNAs in adult zebrafish. Sci Rep 2019;9:3432. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Bartolomé RA, Garcia-Palmero I, Torres S, et al. IL13 Receptor alpha2 Signaling Requires a Scaffold Protein, FAM120A, to Activate the FAK and PI3K Pathways in Colon Cancer Metastasis. Cancer Res 2015;75:2434-44. [Crossref] [PubMed]
- Hung JJ, Jeng WJ, Chou TY, et al. Prognostic value of the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society lung adenocarcinoma classification on death and recurrence in completely resected stage I lung adenocarcinoma. Ann Surg 2013;258:1079-86. [Crossref] [PubMed]
- Cha ST, Chen PS, Johansson G, et al. MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Res 2010;70:2675-85. [Crossref] [PubMed]
- Walsh JC, Lebedev A, Aten E, et al. The clinical importance of assessing tumor hypoxia: relationship of tumor hypoxia to prognosis and therapeutic opportunities. Antioxid Redox Signal 2014;21:1516-54. [Crossref] [PubMed]
- Li S, Li Y, Chen B, et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res 2018;46:D106-12. [Crossref] [PubMed]
- Zhang X, Yang D, Wei Y. Overexpressed CDR1as functions as an oncogene to promote the tumor progression via miR-7 in non-small-cell lung cancer. Onco Targets Ther 2018;11:3979-87. [Crossref] [PubMed]
- Tan S, Gou Q, Pu W, et al. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res 2018;28:693-5. [Crossref] [PubMed]
- Liang G, Liu Z, Tan L, et al. HIF1alpha-associated circDENND4C Promotes Proliferation of Breast Cancer Cells in Hypoxic Environment. Anticancer Res 2017;37:4337-43. [PubMed]
- Boeckel JN, Jae N, Heumuller AW, et al. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ Res 2015;117:884-90. [Crossref] [PubMed]
- Morris LG, Chan TA. Therapeutic targeting of tumor suppressor genes. Cancer 2015;121:1357-68. [Crossref] [PubMed]
- Tanaka M, Sasaki K, Kamata R, et al. A novel RNA-binding protein, Ossa/C9orf10, regulates activity of Src kinases to protect cells from oxidative stress-induced apoptosis. Mol Cell Biol 2009;29:402-13. [Crossref] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]


