The hOGG1 Ser326Cys polymorphism and esophageal cancer risk: a meta-analysis of 1,875 cancer cases and 3,041 controls
Introduction
Esophageal cancer (EC) is the eighth most common cancer worldwide, and the sixth most common cause of cancer-related death (1). Generally, EC has two subtypes, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). ESCC and EAC arise from different cells of the esophagus. The carcinogenesis of EC is still not clear. It is a complex, multi-factorial, and multistep event, in which many factors are involved, such as poor nutritional status, heavy alcohol drinking, cigarette smoking, high-temperature cooking methods, severe lack of vegetable and fruit intake, and genetic factors (2-4). In recent years, genetic factors, including single nucleotide polymorphisms (SNPs), are increasingly regarded as significant contributors to EC (5).
hOGG1, The human 8-oxoguanine glycosylase 1 gene, encodes 8-hydroxygumine DNA glycosylase 1 (OGG1) that can repair damaged DNA by excising 8-dihydro-8-oxoguanine (8-OH-G). Many single nucleotide polymorphisms in this gene are thought to influence the expression of the encoding proteins and activity of encoding proteins, thereby being a predisposition to disease (6). A common single nucleotide polymorphism of hOGG1 at codon 326 polymorphism (Ser326Cys, rs1052133) has been described in recent years. Compared to the 326Ser variant enzyme, the enzyme 326Cys of hOGG1 appears to have a reduced capacity to repair oxidized DNA lesions (7).
Conflicting reports show an association of hOGG1 Ser326Cys polymorphism with different tumor types and ethnic groups (8-13). To date, a considerable number of studies have been conducted to investigate the association between hOGG1 gene polymorphism and EC susceptibility in humans. However, the results remain controversial and ambiguous. In 2013, Zhang et al. (14) conducted a meta-analysis about the association between the hOGG1 Ser326Cys polymorphism and the risk of ESCC, which did not include EAC. In February 2019, Tian et al. (15) collated a comprehensive investigation for cumulative evidence of genetic polymorphisms of EC and its subtype risk. It showed no association between hOGG1 and EC. And the evaluation data were only extracted from previous meta-studies, which may be the main reason for the bias. To derive a more comprehensive and precise estimation of this association, we performed the current meta-analysis.
Methods
Publication search
Computer searches were performed independently by two authors (Chen Zhao and Ji Yang), covering all papers published in PubMed, Embase, Medline, and Google Scholar before November 2018. The keywords were as follows: “EC/oesophageal cancer/ESCC/EAC”, and “OGG1/hOGG1/8-Oxoguanine DNA glycosylase 1”, and “polymorphism/variant/mutation/SNP”. The reference lists of the retrieved articles were hand-searched to obtain other relevant publications. All associated publications were evaluated to identify the most eligible literature. The results were limited to papers published in English. Discrepancies between the two reviewers were resolved by a third reviewer (Liqian Xu).
Inclusion and exclusion criteria
The following criteria were used to select studies for further meta-analysis: (I) the studies were case-controlled; (II) the studies were about hOGG1 Ser326Cys polymorphism and risk of EC; (III) the studies contained at least two comparison groups (cancer group vs. control group); (IV) the studies included detailed genotyping data.
Data extraction
The extraction of data from all eligible publications was performed by two investigators (Chen Zhao and Ji Yang) independently, according to the inclusion and exclusion criteria listed above. Moreover, in cases of conflict, a third reviewer (Liqian Xu) was involved in resolving the conflict. For each study, the information that was extracted was the author’s last name, year of publication, country of origin, ethnicity, cancer type, sources of control and case groups, specimen of cases, genotyping methods for hOGG1 Ser326Cys, total number of cases and controls as well as number of cases and controls with Ser/Ser, Ser/Cys and Cys/Cys genotypes. All of the cases and control groups were well controlled.
Statistical analysis
Hardy-Weinberg equilibrium (HWE) for the control group of each study was assessed using a goodness-of-fit test (χ2 of Fisher’s exact test). Based on both the fixed-effects and random-effects models, and a pooled odds ratio (OR) with 95% confidence interval (CI) was used to assess the strength of association between hOGG1 Ser326Cys polymorphism and EC risk, depending on the heterogeneity of the analysis. In the overall and the subgroup meta-analysis, pooled ORs and 95% CIs for the allele model (Cys vs. Ser) and codominant model (Cys/Cys vs. Ser/Ser; Ser/Cys vs. Ser/Ser); the dominant model (Cys/Cys + Ser/Cys vs. Ser/Ser) and the recessive model (Cys/Cys vs. Ser/Cys + Ser/Ser) were all calculated. Heterogeneity was assessed using Q-test and I2 score. If the results of the heterogeneity test were P>0.05, ORs were pooled according to the fixed-effects model (Mantel-Haenszel model). Otherwise, ORs were pooled according to the random-effects model (DerSimonian and Laird model). I2 was used to qualify variation in OR attributable to heterogeneity.
Publication bias was assessed using the Egger’s test and Begg’s test. All statistical tests were performed using the software STATA v.12.0 (Stata Corporation, College Station, TX, USA). The results were considered statistically significant if P<0.05.
Results
Study selection
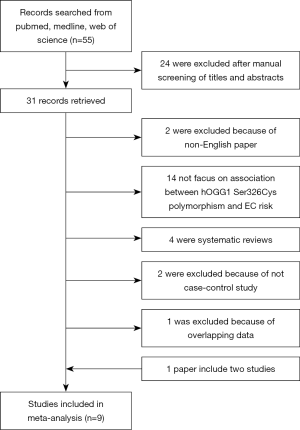
The study selection process is shown in Figure 1. After a preliminary online search, we identified 55 potentially relevant articles for further detailed evaluations. Then, 24 of these were excluded after manual screening of titles and abstracts. Then, a total of 31 records that fulfilled our search criteria were preliminarily identified for further detailed evaluation. Twenty-three studies were excluded because 14 studies were not focused on the association between hOGG1 Ser326Cys polymorphism and EC. Two studies were excluded because they were not written in English. One study was excluded because of overlapping data. Two others were not case-control studies, and the remaining 4 studies were systematic review comments. Finally, 8 records (16-23) on hOGG1 Ser326Cys polymorphism and EC risk were identified, and 1 of them included 2 studies. Finally, a total of 9 case-control studies involving 1,875 cancer cases and 3,041 controls were included in the meta-analysis.
Study characteristics
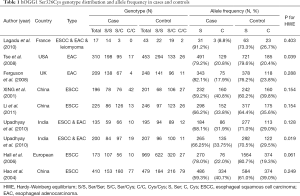
Characteristics of the papers included in this meta-analysis are presented in Table 1. All studies involved in published papers are case-control studies. The studies were carried out in China, UK, France, USA, European, and India. The studies carried out in China and India were used in the Asian subgroup, and the others were used in the Caucasian subgroup. Of the 9 studies, we found that 2 were related to EAC, 4 were related to ESCC, and another 3 studies contained both types of EC (EAC and ESCC) but we could not obtain the specific figures of EAC/ESCC patients even after contacting the authors. The genotype distribution in the controls was consistent with the HWE (P>0.05) in other studies except for two studies (Tse et al., P=0.039; Upadhyay et al., P=0.019).
Full table
Quantitative data synthesis
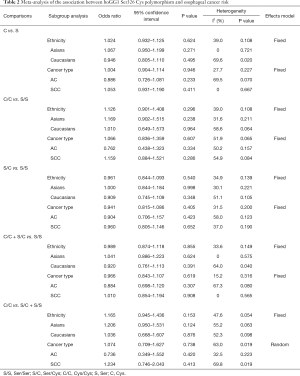
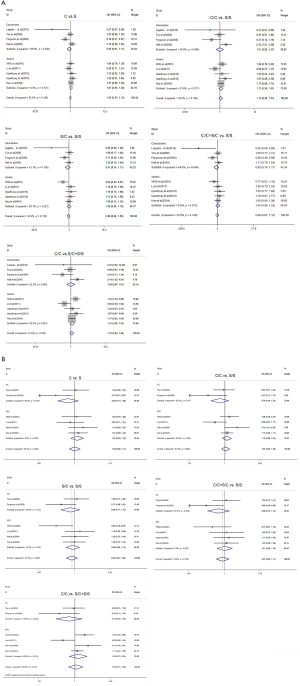
The results on the associations between hOGG1 Ser326Cys polymorphism and EC risk, and of the heterogeneity test, are shown in Table 2. Overall, when all of the eligible studies were pooled into the meta-analysis, no significant associations between hOGG1 Ser326Cys polymorphism and EC susceptibility were found for the allele model: Cys vs. Ser (OR =1.024, 95% CI: 0.932–1.125, P=0.624); codominant model: Cys/Cys vs. Ser/Ser (OR =1.126, 95% CI: 0.901–1.408, P=0.296), Ser/Cys vs. Ser/Ser(OR =0.961, 95% CI: 0.844–1.093, P=0.540); dominant model: Cys/Cys + Ser/Cys vs. Ser/Ser (OR =0.989, 95% CI: 0.874–1.118, P=0.855); recessive model: Cys/Cys vs. Ser/Cys + Ser/Ser (OR =1.165, 95% CI: 0.945–1.436, P=0.153) (Figure 2). In the stratified analysis by ethnicity, no significant results were found for Asian and Caucasian subjects in different statistical models (all P>0.05) (Table 2, Figure 2A). Similarly, no significant difference was observed in different cancer types (all P>0.05) (Table 2, Figure 2B).
Full table
Tests of heterogeneity
No statistically significant heterogeneity was observed between trials of the following analyses using Q statistic and I2 score (Cys vs. Ser: P=0.108, I2=39.0%; Cys/Cys vs. Ser/Ser: P=0.108, I2=39.0%; Ser/Cys vs. Ser/Ser: P=0.139, I2=34.9%; Cys/Cys + Ser/Cys vs. Ser/Ser: P=0.149, I2=33.6%. Cys/Cys vs. Ser/Cys + Ser/Ser: P=0.054, I2=47.6%) (Table 2), and the fixed-effects model was employed in these studies.
Publication bias
Egger’s test and Begg’s test were performed to assess publication bias. The Egger weighted regression method did not indicate any evidence for publication bias (Cys vs. Ser: P=0.106; Cys/Cys vs. Ser/Ser: P=0.452; Ser/Cys vs. Ser/Ser: P=0.101; Cys/Cys + Ser/Cys vs. Ser/Ser: P=0.086; Cys/Cys vs. Ser/Cys + Ser/Ser: P=0.526). This result was confirmed by the Begg rank correlation method (Cys vs. Ser: P=0.754; Cys/Cys vs. Ser/Ser: P=0.602; Ser/Cys vs. Ser/Ser: P=0.754; Cys/Cys + Ser/Cys vs. Ser/Ser: P=0.466; Cys/Cys vs. Ser/Cys + Ser/Ser: P = 0.602) (Table 3).

Full table
Discussion
EC is a malignant tumor of the esophagus, and the mechanisms for EC remains unclear. Several studies have found that DNA repair efficiency is lower in cancer patients than in that of normal people, and the variants of the genes involved in DNA repair can increase the cancer risk (24). Among many factors of oxidative DNA damage, 8-hydroy-2-deoxyguanine (8-OHdG) is one of the most abundant oxidative products of highly mutagenic because of its propensity to mispair with adenine during DNA replication (25). Studies have shown that the hOGG1 gene could remove 8-OHdG from DNA by base excision repair (BER) pathway (6,26). Another report also revealed that genetic variations in hOGG1 gene might alter glycosylase activity, increasing the cancer risk (27). There are several polymorphisms in the hOGG1 gene (28), and Ser326Cys polymorphism has attracted widespread attention.
Many epidemiological studies have investigated the association of the Ser326Cys polymorphism in the hOGG1 gene with different types of cancers, but there are conflicting reports. Also, studies on the prevalence of this polymorphism in susceptibility to EC show conflicting results. Xing et al. (17) reported that homozygosity for the Cys/Cys genotype significantly increased the risk of developing ESCC in Asians, which indicated that the hOGG1 326Cys allele might play a role in the carcinogenesis of the esophagus whereas Tse et al. (18) and Ferguson et al. (19) reported no statistical association between hOGG1 gene polymorphism and EAC in Caucasians. Hence, this meta-analysis was needed to provide a quantitative approach for combining the different results. A study conducted in Asians by Upadhyay et al. (21) indicated that no significant association was found between hOGG1 Ser326Cys genotypes and EC risk. To derive a more precise estimation of the association, we conducted a meta-analysis. The present meta-analysis, including 1,875 cancer cases and 3,041 controls, explored the relationship between the hOGG1 Ser326Cys polymorphism and the overall EC risk. This meta-analysis on hOGG1 Ser326Cys polymorphism and the risk of EC suggests no statistically significant association between the two. Additional primary studies may be necessary to provide evidence of any significant association between this specific polymorphism and EC. We found that there was no statistically significant difference between hOGG1 Ser326Cys polymorphism and EC risk in any types of the statistical model (all P>0.05). In the subgroup meta-analysis based on ethnicity, no significant results were found for Asian and Caucasian subjects in the different statistical model (all P>0.05). Similarly, no significant results were found for ESCC and EAC in the different statistical model (all P>0.05). These results were inconsistent with those of Zhang et al. A meta-analysis conducted by Zhang et al. in 2013 (14) suggested that the hOGG1 Ser326Cys polymorphism was associated with ESCC susceptibility. Cys/Cys carriers have more risk on ESCC rather than Ser/Ser and Ser/Cys carriers. The reasons for this difference were as follows: firstly, the literature we selected was limited to English, which limited the scale of the data to the meta-analysis and avoided duplication of studies enrolled; secondly, two studies enrolled by Zhang et al. were abandoned when we read the full text of articles because they are mixed cancer types (ESCC and EAC). These results revealed that ethnicity or environment might not be critical factors on the effects of the polymorphic alleles. Further studies are still needed to use standardized unbiased homogenous cancer patients and well-matched controls to investigate the combined effects.
Several limitations of this meta-analysis should be addressed. First, the analysis did not consider gene-gene and gene-environment interactions, and a more precise analysis might be conducted, which could allow for an adjustment estimate by sex, age, and lifestyle such as smoking and alcohol drinking. Second, the controls were not uniformly defined, as although most of the patients in the control groups were selected from healthy populations, some might have benign disease. Third, they were mixed cancer types, and we could not get the numbers of ESCC or EAC patients from studies or authors, which may limit the scale of the data in the meta-analysis.
In conclusion, the current meta-analysis suggests that hOGG1 Ser326Cys polymorphism is the lack of any association with EC risk. Our results should be interpreted with cautions considering the limited studies in both overall and subgroup analyses. Unbiased, well designed, and prospective studies with a larger sample size focusing on diverse ethnicities, sex, age, lifestyle, and pathological cancer types should be conducted to determine further whether there exist any correlations between the hOGG1 Ser326Cys polymorphism and the risk of EC.
Acknowledgments
We thank Yingyu Ma (Key Laboratory of Gastroenterology of Zhejiang Province, Zhejiang Provincial People’s Hospital) for her help and support to the methodology and meta process.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was a systematic review and meta-analysis of all published literature and institutional review deemed this study exempt from ethical approval.
References
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893-917. [Crossref] [PubMed]
- Yu MC, Garabrant DH, Peters JM, et al. Tobacco, alcohol, diet, occupation, and carcinoma of the esophagus. Cancer Res 1988;48:3843-8. [PubMed]
- Farrow DC, Vaughan TL, Sweeney C, et al. Gastroesophageal reflux disease, use of H2 receptor antagonists, and risk of esophageal and gastric cancer. Cancer Causes Control 2000;11:231-8. [Crossref] [PubMed]
- Lin J, Zeng R, Cao W, et al. Hot beverage and food intake and esophageal cancer in southern China. Asian Pac J Cancer Prev 2011;12:2189-92. [PubMed]
- Wijnhoven BP, Toxopeus EL, Vallböhmer D, et al. New therapeutic strategies for squamous cell cancer and adenocarcinoma. Ann N Y Acad Sci 2013;1300:213-25. [Crossref] [PubMed]
- Boiteux S, Radicella JP. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch Biochem Biophys 2000;377:1-8. [Crossref] [PubMed]
- Kohno T, Shinmura K, Tosaka M, et al. Genetic polymorphisms and alternative splicing of the hOGG1 gene, that is involved in the repair of 8-hydroxyguanine in damaged DNA. Oncogene 1998;16:3219-25. [Crossref] [PubMed]
- Michalska MM, Samulak D, Romanowicz H, et al. Single nucleotide polymorphisms (SNPs) of hOGG1 and XRCC1 DNA repair genes and the risk of ovarian cancer in Polish women. Tumour Biol 2015;36:9457-63. [Crossref] [PubMed]
- Kamali M, Kargar S, Heiranizadeh N, et al. Lack of any Association between the Hogg1 Ser326Cys Polymorphism and Breast Cancer Risk: a Systematic Review And Meta-Analysis Of 18 Studies. Asian Pac J Cancer Prev 2017;18:245-51. [PubMed]
- Wenjuan C, Jianzhong L, Chong L, et al. The hOGG1 Ser326Cys gene polymorphism and susceptibility for bladder cancer: a meta-analysis. Int Braz J Urol 2016;42:883-96. [Crossref] [PubMed]
- Hu WG, Pan RJ, Cai W, et al. Lack of association between the hOGG1 gene Ser326Cys polymorphism and gastric cancer risk: evidence from a case-control study and a meta-analysis. Genet Mol Res 2015;14:14670-9. [Crossref] [PubMed]
- Guo J, Yang J, Li Y. Association of hOGG1 Ser326Cys polymorphism with susceptibility to hepatocellular carcinoma. Int J Clin Exp Med 2015;8:8977-85. [PubMed]
- Marques CR, Da Silva TM, De Albuquerque DM, et al. NAT2, XRCC1 and hOGG1 polymorphisms, cigarette smoking, alcohol consumption and risk of upper aerodigestive tract cancer. Anticancer Res 2014;34:3217-24. [PubMed]
- Zhang J, Zhou J, Zhang P, et al. A meta-analysis of the association between the hOGG1 Ser326Cys polymorphism and the risk of esophageal squamous cell carcinoma. PLoS One 2013;8:e65742. [Crossref] [PubMed]
- Tian J, Liu C, Liu G, et al. Cumulative evidence for association between genetic polymorphisms and esophageal cancer susceptibility: A review with evidence from meta-analysis and genome-wide association studies. Cancer Med 2019;8:1289-305. [Crossref] [PubMed]
- Lagadu S, Lechevrel M, Sichel F, et al. 8-oxo-7,8-dihydro-2’-deoxyguanosine as a biomarker of oxidative damage in oesophageal cancer patients: lack of association with antioxidant vitamins and polymorphism of hOGG1 and GST. J Exp Clin Cancer Res 2010;29:157. [Crossref] [PubMed]
- Xing DY, Tan W, Song N, et al. Ser326Cys polymorphism in hOGG1 gene and risk of esophageal cancer in a Chinese population. Int J Cancer 2001;95:140-3. [Crossref] [PubMed]
- Tse D, Zhai R, Zhou W, et al. Polymorphisms of the NER pathway genes, ERCC1 and XPD are associated with esophageal adenocarcinoma risk. Cancer Causes Control 2008;19:1077-83. [Crossref] [PubMed]
- Ferguson HR, Wild CP, Anderson LA, et al. No association between hOGG1, XRCC1, and XPD polymorphisms and risk of reflux esophagitis, Barrett’s esophagus, or esophageal adenocarcinoma: results from the factors influencing the Barrett’s adenocarcinoma relationship case-control study. Cancer Epidemiol Biomarkers Prev 2008;17:736-9. [Crossref] [PubMed]
- Li QD, Li H, Wang MS, et al. Multi-susceptibility genes associated with the risk of the development stages of esophageal squamous cell cancer in Feicheng County. BMC Gastroenterol 2011;11:74. [Crossref] [PubMed]
- Upadhyay R, Malik MA, Zargar SA, et al. OGG1 Ser326Cys polymorphism and susceptibility to esophageal cancer in low and high at-risk populations of northern India. J Gastrointest Cancer 2010;41:110-5. [Crossref] [PubMed]
- Hall J, Hashibe M, Boffetta P, et al. The association of sequence variants in DNA repair and cell cycle genes with cancers of the upper aerodigestive tract. Carcinogenesis 2007;28:665-71. [Crossref] [PubMed]
- Hao B, Wang H, Zhou K, et al. Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer Res 2004;64:4378-84. [Crossref] [PubMed]
- Mohrenweiser HW, Wilson DM, Jones IM. Challenges and complexities in estimating both the functional impact and the disease risk associated with the extensive genetic variation in human DNA repair genes. Mutat Res 2003;526:93-125. [Crossref] [PubMed]
- Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 1991;349:431-4. [Crossref] [PubMed]
- Boiteux S, Radicella JP. Base excision repair of 8-hydroxyguanine protects DNA from endogenous oxidative stress. Biochimie 1999;81:59-67. [Crossref] [PubMed]
- Audebert M, Radicella JP, Dizdaroglu M. Effect of single mutations in the OGG1 gene found in human tumors on the substrate specificity of the Ogg1 protein. Nucleic Acids Res 2000;28:2672-8. [Crossref] [PubMed]
- Nishioka K, Ohtsubo T, Oda H, et al. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol Biol Cell 1999;10:1637-52. [Crossref] [PubMed]