
Identification of CD2, CCL5 and CCR5 as potential therapeutic target genes for renal interstitial fibrosis
Introduction
Most chronic kidney disease (CKD) would finally turn into End-Stage Renal Disease (ESRD), impacting 12% of all adults in the United States and 10.8% in China (1). By 2030, the number of ESRD patients who need maintenance hemodialysis would increase to about 5.4 billion (2). Currently, renal transplantation is recognized as the best treatment for ESRD as life expectancy in renal transplant patients has improved significantly (3). Over the past 20 years, although the success rate and 1-year survival rate of renal transplantation has improved, long-term survival rate remained dissatisfied. By report, long-term survival rate was 68% for living-related renal transplantation and 51% for cadaveric renal transplantation (4).
Renal interstitial fibrosis (RIF) of the transplanted renal, as a pathophysiological process, acts as the main pathological feature of renal transplantation failure, whose clinical manifestation was the decrease of glomerular filtration rate (5). RIF is related to antibody-mediated rejection (AMR) (6), epithelial to mesenchymal transition (EMT) (7) and excessive accumulation of extracellular matrix (ECM) (8). Presently, examination-renal biopsy is still the gold standard for the diagnosis of RIF. However, as an invasive method, biopsy difficulty, biopsy-sampling error and diagnostic deviation of pathologist may affect diagnosis results, which limits its application. Recently, some serum biomarkers including urinary-specific matrix metalloproteinase (MMP)-generated collagen type I and III degradation fragments (9) might help predict the progression of RIF. However, the specificity of serum biomarkers is still unsatisfied due to interference of other renal diseases such as glomerular nephritis and acute kidney injury. Therefore, it is particularly important to find effective and non-invasive methods for early diagnosis of RIF, which could help adjust the therapeutic regimen timely so as to improve the function of transplanted renal.
Recently, mRNAs have raised great interest to be detected as biomarkers for RIF. Cao et al. pointed that vimentin (VIM) Mrna presented the highest change fold of 9.99 in RIF patients, indicating it might be assumed as a novel non-invasive biomarker to monitor the process of RIF (10). Moreover, studies indicated that RIF was associated with immune inflammatory signaling pathways such as TGF-β/Smad (11) and Wnt/β-catenin (12) pathway. However, limited mechanism of RIF has been discovered. Accordingly, we aimed to identify the differentially expressed genes (DEGs) using the Gene Expression Omnibus (GEO) database. Then we conducted Gene Ontology (GO) function enrichment analysis and protein-protein interaction (PPI) network analysis to excavate the potential biological action mechanisms and hub genes for further exploration.
Methods
Data acquisition
The data of RIF and normal samples were derived from the GEO database. The result was shown in Table 1. There were 24 interstitial fibrosis and tubular atrophy (IFTA) and 16 interstitial fibrosis and tubular atrophy with inflammation (IFTA_i) samples in the GSE22459 data set; 43 IFTA samples, 10 IFTA_i and 54 interstitial fibrosis and tubular atrophy with acute rejection (IFTA_AR) samples in the GSE76882 data set. They were then applied to identify the DEGs. The GSE57731 data set (including 45 normal samples and 14 IFTA samples) was mainly employed to verify the relationship between hub gene and the stage of RIF.
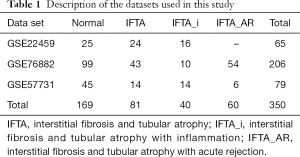
Full table
Data pretreatment
Raw data was downloaded from GEO database (CEL files). We used Bioconductor ‘limma’ package to perform background adjustment. Through the platform files and the Chip signal data which comes from gene chip, gene (Gene Symbol) and probe identification (ID) were selected. If a probe set was corresponded to multiple or 0 genes, the data of that probe set was omitted. If multiple probe sets were mapped to the same gene, the gene expression profile data was calculated by the arithmetic mean value of multiple probe sets.
Identification of DEGs
Cluster analysis was conducted to explore the change of gene expression patterns between IFTA samples and normal samples. It was carried out by ‘edgeR’ package in R software (13). |log2 fold-change| >2 and the P value of false discovery rate (FDR) <0.05 was settled as a threshold (14). Through application of Jaccard distance and complete linkage, we analyzed the clustering and formed clusters. Significance analysis of microarrays was applied to identify the DEGs. Besides, we generated heat maps using ‘pheatmap’ package in R software.
Function enrichment
GO enrichment analysis was then conducted by DAVID (database for annotation, visualization, and integrated discovery) (15) with FDR <0.01 to screen significant functional pathway enriched in the DEGs. We applied GO plot package of R software to explore the GO terms.
Establishment of PPI network
For the purpose of verifying the potential relationships of DEGs, STRING (Search Tool for the Retrieval of Interacting Genes) database (16) was employed to construct the PPI network to find hub genes. The modification and visualization of network was performed by Cytoscape 3.6.1 based on the JAVA software (17).
Correlation analysis
We used Graphpad Prism 7 to calculate the Spearman’s rank correlation coefficient to explore the correlation between three hub genes (CD2, CCL5 and CCR5) and pathological degrees of RIF.
Results
DEG patterns
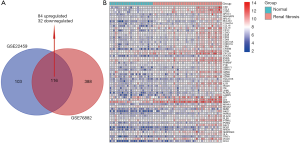
To investigate the differences in gene expression between IFTA samples and normal samples, we used the sample set GSE22459 which contained 25 normal samples, 24 IFTA samples and 16 IFTA_i samples to identify 219 DEGs. Among them, 55 genes were lower expressed, while 164 genes were higher expressed of IFTA samples. At the same time, another sample set GSE76882 which included 99 normal samples, 43 IFTA samples, 10 IFTA_i samples and 54 IFTA_AR samples was used to verify 504 DEGs (184 was downregulated and 320 was upregulated). Next, 116 overlapping genes (84 up-regulated and 32 down-regulated) were obtained by intersecting the two sets of DEGs (Figure 1A). Then we performed cluster analysis for all 116 DEGs. Only the top 60 genes with the most significant differences were selected for visualization in Figure 1B. It suggested that these genes had obvious different gene expression patterns between normal renal samples and IFTA samples.
Function annotation of DEGs
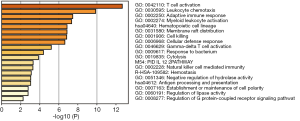
GO analysis was conducted to obtain the biological functions of 116 DEGs to understand which signaling pathways might serve as an important role in RIF. As shown in Figure 2, results showed that these DEGs were enriched in 19 GO biological process categories. Some pathways were related to inflammation and immunity such as T cell activation, antigen processing and presentation, natural killer cell mediated immunity, myeloid leukocyte activation, adaptive immune response and leukocyte chemotaxis, implying that there might be abnormal inflammatory and immune responses in IFTA samples.
Identification of hub genes via PPI network
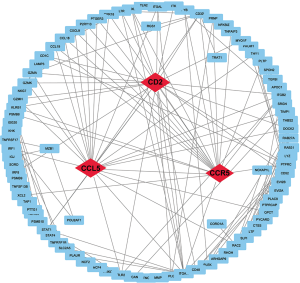
We constructed a PPI network to investigate the underlying relationships among these genes, thus detecting 97 nodes and 143 edges. What’s more, Cytoscape 3.6.1 was applied to screen hub genes which were pivotal nodes in this network. As shown in Figure 3, CD2, CCL5 and CCR5 acted as the main hub genes.
Three hub genes (CD2, CCL5 and CCR5) correlated with pathological degrees of RIF
We selected the independent sample set GSE57731 from the GEO database to further explore the relationship between hub genes with the pathological degree of RIF. Since most RIF events occurred in the first 3 months after renal transplantation, we used the GSE57731 sample set which came from the blood samples collected in 3 months. CD2, CCL5 and CCR5 were found to kept higher expression patterns in high-fibrosis samples. Moreover, their gene expression levels were found significantly positive correlation with the degree of RIF (Figure 4, CD2: P<0.05, r=0.29; CCL5: P<0.05, r=0.31; CCR5: P<0.05, r=0.38).

Discussion
Renal transplantation is the most mature and reliable method for the treatment of ESRD, which could effectively improve the quality of life and survival time of patients (18). However, the long-term survival of allogeneic renal transplantation remained unsatisfactory (19). A cohort study illustrated that RIF was an important time-dependent histologic lesion, occurring in 70% of biopsy samples in 3 months on post-transplantation (20). Immune cells (macrophages, T lymphocytes and mast cells) (21) and profibrogenic cytokines (TGF-1 and CTGF) (22) were involved in the occurrence and development of RIF. Screening the early potential targets for diagnosis was necessary as it was reported that early graft function held a significant influence on long-term renal function (23).
Nowadays, both diagnosis and intervention of RIF were challenges for clinicians. Glomerular filtration rate and serum creatinine clearance had limited clinical utility in assessing renal histopathological changes and fibrosis after renal transplantation. In addition, the non-invasive diagnosis of RIF was tough with the interference of complex immune regulatory network and non-immune factors (24). As serum markers were easily influenced by other renal diseases, most recent discoveries of diagnostic biomarkers have arisen in the field of genetic molecules such as microRNA (miRNA). For example, MiR-433 was an important component of TGF-β/Smad3 pathways and could enhance TGF-β1-induced fibrosis by enhancing the antizyme inhibitor—Azin1 in vitro and in vivo (25). Jenkins et al. found that miR-192 mediated the development of tubulointerstitial fibrosis via repression of E-box repressors such as ZEB1, causing an activation of akt kinase signaling to increase the expression of collagen (26).
In this study, we identified 116 DEGs between IFTA samples and normal samples based on data sets downloaded from GEO database. All samples were obtained directly from fibrous renal tissue to improve their accuracy. Among these DEGs, GO analysis was conducted to verify some pathways of inflammation and immunity such as T cell activation and natural killer cell mediated immunity. Finally, we detected three DEGs (CD2, CCL5 and CCR5) which may be the potential molecular mechanisms and biomarkers by performing PPI network.
DEGs were found enriched in some pathways related to the inflammation and immunity. “T cell activation” was the most enriched GO terms in BP. A study suggested that the recruitment and activation of CD4+ T lymphocyte could be the critical step of the onset of RIF so that the suppression of CD4+ T lymphocyte differentiation to Th2 could be a potential therapeutic intervention (27). Besides, ‘Natural killer cell mediated immunity’ term was also enriched. NK cells were subsets of lymphocytes and played an important role in regulating the immune response. Law et al. found that the number of total NK cells (CD3−CD56+) were detected higher in RIF tissues compared with tissues without fibrosis, indicating NK cells was a vital factor to fibrotic process (28). Other certified immune-related terms included antigen processing and presentation, myeloid leukocyte activation, adaptive immune response and leukocyte chemotaxis.
CD2 which located in 1p13.1 is highly expressed in RIF. Its encoding protein is a surface antigen found on all peripheral blood T-cells. Moreover, CD2-positive T cells were the predominant infiltrating cells in various glomerulonephritis (29). Relevant studies on CD2AP encoded by CD2 have been reported that CD2AP was also involved in a variety of signaling pathways. By selectively enhancing the expression of TGF-β, CD2AP could activate the TGF-β/smad3 signaling pathway which promoted the occurrence of RIF (30). What’s more, CD2AP was important for maintaining the integrity of slit diaphragm and cytoskeleton (31). Elevated CD2 expression might be a compensatory repair mechanism after renal tissue injury. Besides, the reduction of urinary exosome mRNA of CD2AP in RIF made it possible to be a noninvasive detection biomarker of renal fibrosis (32).
CCL5 was one of several chemokine genes clustered on the q-arm of chromosome 17. In renal transplantation rejection, molecular biology studies have confirmed that the activation of NF-κB signaling enhanced the secretion of TGF-β, upregulating CCL5 in mesangial cells to promote RIF (33). Furthermore, CCL5 used chemokine receptor chemokine (C-C motif) receptor 5 (CCR5) that was found to be highly expressed in our study as a co-receptor (34). CCR5 encoded a member of the beta chemokine receptor family, which was predicted to be a seven transmembrane protein similar to G protein-coupled receptors. A study pointed that inhibition of a potent mitogen for fibroblasts PDGF-C produced a decrease in the expression of the renal chemokines CCL5, accompanied by a significant reduction in expression of CCR mRNA. We hypothesized that CCL5 could be combined with CCR5+ T cells to promote the aggregation of monocytes or macrophages, thus leading to the occurrence of immune inflammatory response. However, further exploration is needed (35). Moreover, research has shown that CCR5 may be a potential drug target for anti-fibrosis therapy (36).
Admittedly, we found that there were significantly different gene patterns between high- and low-fibrosis samples. GSE57731 which was a serological sample was employed to verify it. It was cheaper and safer to get the patients’ serum than to obtain their kidney tissue with less damage to the patients. Gene expression levels of them were remarkably positively correlated with the degree of RIF, indicating that there were changes in expression of these genes in the early stages of disease. Research has reported that the expression of CCL5 in RIF increased with the increase of pathological grading (37). As early graft function held a significant effect on long-term renal function (23), CD2, CCL5 and CCR5 have potential to be the early biomarkers of diagnosis. If the biomarkers acquired from fibrous renal tissue could be verified in serological samples, it would be of great value of medical application and research.
The use of clinical biomarkers is easier and less expensive than direct measurement of the final clinical endpoint. To be honest, there existed some studies which focused on the specific role of these genes in the mechanism of renal fibrosis related to these three genes. It was necessary, but was just the first step to know the pathophysiological relationship between the biomarker and the relevant clinical endpoint. In this research, we emphasized on transforming these basic studies into clinical applications. We hoped that they can be used in screening, diagnosis, characterization and monitoring of RIF.
Our study still has some limitations. First, we were currently unable to conduct the multi-center clinical verification. Second, due to the limited research conditions, we could not perform in vitro or in vivo experiments presently. So, we have limited evidence to explain the mechanism of RIF. Nevertheless, we found three DEGs (CD2, CCL5 and CCR5) which may be the potential molecular mechanisms and markers for further study.
Acknowledgments
Funding: This work was supported by grants from Excellent Young Talents in Clinical Medicine of Hongkou District (HKYQ2018-09), the National Natural Science Foundation of China (No. 81702520), Medical Research Project of Jiangsu Provincial Health and Family Planning Commission (No. H2018052), Research Project of Jiangsu Cancer Hospital (No. ZN201602), and the young talents program of Jiangsu Cancer Hospital (No. 2017YQL-04).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Jha V, Garcia-Garcia G, Iseki K, et al. Chronic kidney disease: global dimension and perspectives. Lancet 2013;382:260-72. [Crossref] [PubMed]
- Castro A, Neri M, Karopadi AN, et al. How can we advance in renal replacement therapy techniques? Nefrologia 2019;39:372-8. [Crossref] [PubMed]
- Long B, Koyfman A, Lee CM. Emergency medicine evaluation and management of the end stage renal disease patient. Am J Emerg Med 2017;35:1946-55. [Crossref] [PubMed]
- Lamb KE, Lodhi S, Meier-Kriesche HU. Long-term renal allograft survival in the United States: a critical reappraisal. Am J Transplant 2011;11:450-62. [Crossref] [PubMed]
- Nogueira A, Pires MJ, Oliveira PA. Pathophysiological Mechanisms of Renal Fibrosis: A Review of Animal Models and Therapeutic Strategies. In Vivo 2017;31:1-22. [Crossref] [PubMed]
- Sellares J, de Freitas DG, Mengel M, et al. Understanding the causes of kidney transplant failure: the dominant role of antibody-mediated rejection and nonadherence. Am J Transplant 2012;12:388-99. [Crossref] [PubMed]
- Cruz-Solbes AS, Youker K. Epithelial to Mesenchymal Transition (EMT) and Endothelial to Mesenchymal Transition (EndMT): Role and Implications in Kidney Fibrosis. Results Probl Cell Differ 2017;60:345-72. [Crossref] [PubMed]
- Klinkhammer BM, Goldschmeding R, Floege J, et al. Treatment of Renal Fibrosis-Turning Challenges into Opportunities. Adv Chronic Kidney Dis 2017;24:117-29. [Crossref] [PubMed]
- Papasotiriou M, Genovese F, Klinkhammer BM, et al. Serum and urine markers of collagen degradation reflect renal fibrosis in experimental kidney diseases. Nephrol Dial Transplant 2015;30:1112-21. [Crossref] [PubMed]
- Cao YH, Lv LL, Zhang X, et al. Urinary vimentin mRNA as a potential novel biomarker of renal fibrosis. Am J Physiol Renal Physiol 2015;309:F514-22. [Crossref] [PubMed]
- Li H, Cai H, Deng J, et al. TGF-beta-mediated upregulation of Sox9 in fibroblast promotes renal fibrosis. Biochim Biophys Acta Mol Basis Dis 2018;1864:520-32.
- Lin X, Zha Y, Zeng XZ, et al. Role of the Wnt/beta-catenin signaling pathway in inducing apoptosis and renal fibrosis in 5/6-nephrectomized rats. Mol Med Rep 2017;15:3575-82. [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [Crossref] [PubMed]
- Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:3. [Crossref] [PubMed]
- Liu Y, Zhang M, Liang L, et al. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol 2015;8:11480-4. [PubMed]
- Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 2010;17:1471-4.
- Broers NJH, Fung TY, Kooman JP, et al. Living-donor transplantation leads to a major improvement in physical functioning: an observational study on the impact on potential donors and their recipients. BMC Nephrol 2019;20:109. [Crossref] [PubMed]
- San Segundo D, Galvan-Espinoza LH, Rodrigo E, et al. Regulatory T-cell Number in Peripheral Blood at 1 Year Posttransplant as Predictor of Long-term Kidney Graft Survival. Transplant Direct 2019;5:e426. [Crossref] [PubMed]
- Nankivell BJ, Shingde M, Keung KL, et al. The causes, significance and consequences of inflammatory fibrosis in kidney transplantation: The Banff i-IFTA lesion. Am J Transplant 2018;18:364-76. [Crossref] [PubMed]
- Lawson J, Elliott J, Wheeler-Jones C, et al. Renal fibrosis in feline chronic kidney disease: known mediators and mechanisms of injury. Vet J 2015;203:18-26. [Crossref] [PubMed]
- Xavier S, Vasko R, Matsumoto K, et al. Curtailing endothelial TGF-beta signaling is sufficient to reduce endothelial-mesenchymal transition and fibrosis in CKD. J Am Soc Nephrol 2015;26:817-29. [Crossref] [PubMed]
- Pita-Fernandez S, Valdes-Canedo F, Seoane-Pillado T, et al. Influence of early graft function after renal transplantation and its impact on long-term graft and patient survival. Transplant Proc 2010;42:2856-8. [Crossref] [PubMed]
- Zununi Vahed S, Samadi N, Ardalan M. Diagnosis of interstitial fibrosis and tubular atrophy in kidney allograft: implementation of microRNAs. Iran J Kidney Dis 2014;8:4-12. [PubMed]
- Li R, Chung AC, Dong Y, et al. The microRNA miR-433 promotes renal fibrosis by amplifying the TGF-beta/Smad3-Azin1 pathway. Kidney Int 2013;84:1129-44. [Crossref] [PubMed]
- Jenkins RH, Martin J, Phillips AO, et al. Pleiotropy of microRNA-192 in the kidney. Biochem Soc Trans 2012;40:762-7. [Crossref] [PubMed]
- Liu L, Kou P, Zeng Q, et al. CD4+ T Lymphocytes, especially Th2 cells, contribute to the progress of renal fibrosis. Am J Nephrol 2012;36:386-96. [Crossref] [PubMed]
- Law BM, Wilkinson R, Wang X, et al. Interferon-gamma production by tubulointerstitial human CD56(bright) natural killer cells contributes to renal fibrosis and chronic kidney disease progression. Kidney Int 2017;92:79-88. [Crossref] [PubMed]
- Floege J, Amann K. Primary glomerulonephritides. Lancet 2016;387:2036-48. [Crossref] [PubMed]
- Samah M, El-Aidy Ael R, Tawfik MK, et al. Evaluation of the antifibrotic effect of fenofibrate and rosiglitazone on bleomycin-induced pulmonary fibrosis in rats. Eur J Pharmacol 2012;689:186-93. [Crossref] [PubMed]
- Tienari J, Lehtonen S, Lehtonen E. CD2-associated protein in human urogenital system and in adult kidney tumours. Virchows Arch 2005;446:394-401. [Crossref] [PubMed]
- Lv LL, Cao YH, Pan MM, et al. CD2AP mRNA in urinary exosome as biomarker of kidney disease. Clin Chim Acta 2014;428:26-31. [Crossref] [PubMed]
- Schmid H, Boucherot A, Yasuda Y, et al. Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes 2006;55:2993-3003. [Crossref] [PubMed]
- Oppermann M. Chemokine receptor CCR5: insights into structure, function, and regulation. Cell Signal 2004;16:1201-10. [Crossref] [PubMed]
- Eitner F, Bucher E, van Roeyen C, et al. PDGF-C is a proinflammatory cytokine that mediates renal interstitial fibrosis. J Am Soc Nephrol 2008;19:281-9. [Crossref] [PubMed]
- Furuichi K, Wada T, Sakai N, et al. Distinct expression of CCR1 and CCR5 in glomerular and interstitial lesions of human glomerular diseases. Am J Nephrol 2000;20:291-9. [Crossref] [PubMed]
- Newcomb B, Rhein C, Mileva I, et al. Identification of an acid sphingomyelinase ceramide kinase pathway in the regulation of the chemokine CCL5. J Lipid Res 2018;59:1219-29. [Crossref] [PubMed]


