
Precision oncology for gallbladder cancer: insights from genetic alterations and clinical practice
Introduction
Gallbladder cancer (GBC) is a relatively uncommon aggressive neoplasm with a poor prognosis. GBC is more prevalent in Chile and India than in other countries (1). Patients with GBC often experience disease progression or recurrence shortly after surgery, and the availability of adjuvant therapies for GBC is limited (2). For systemic chemotherapy, only the gemcitabine plus cisplatin regimen has been was approved for the patients with advanced biliary tract cancers (3). Although numerous trials have attempted to test the efficacy of targeted drugs administered in monotherapy or combination treatments, no targeted therapeutic regimen has been approved for the treatment of advanced GBC (4). The shortage of available targeted therapies is potentially the result of the complex biological characteristics of GBC and the lack of insight into the genomic landscape of this rare type of tumor.
Genomic profiling-guided targeted therapy (GPTT) has been widely applied in several types of cancers, which has promoted the development of research to identify the diverse mutations in the cancer genome (5). Preclinical studies focused on the genome of biliary tract cancers have identified its heterogeneous mutations and molecular alterations (6). Cancer-associated mutations that occur with a high frequency in GBC have been identified, and ongoing trials targeting these mutations have shown promising outcomes (7). Trials targeting EGFR, IDH1/2, ERBB2 and FGFR have recently demonstrated encouraging outcomes for patients with advanced biliary tract cancers (8-10). However, the proportion of GBC patients obtaining clinical benefits from these therapies is still limited, likely due to the lack of translational research available to fill the gap between the genomic alteration spectra and the management of clinical therapy.
We formed a multidisciplinary tumor board (MDTB) to conduct an open-label, single-center, prospective clinical trial to assess the feasibility of GPTT among Chinese patients with advanced hepatobiliary cancer (Figure 1). The present study primarily aims to explore the landscape of genomic alterations in GBC and to further identify potentially actionable targets.
Methods
Study design
This was a proof-of-concept, open-label, single-center, prospective clinical trial to determine the mutational spectra in Chinese GBC patients and to assess the feasibility of GPTT in GBC patients through real-world clinical practice (PTHBC, NCT02715089). The therapeutic decision for each participant was discussed and formulated by a MDTB in Department of Liver Surgery from Peking Union Medical College Hospital (PUMCH). The primary purpose of this pilot study was to explore the frequency of actionable genomic alterations detected by high-throughput sequencing in patients with advanced GBC, and to determine the feasibility of individualized cancer management based on GPTT. This trial was approved by the local ethics committee of PUMCH. All participants provided written informed consent.
Patient population
The main inclusion criteria were as follows: (I) males or females aged 18 to 75 years; (II) pathologically confirmed stage III or IV GBC; (III) advanced stage GBC with recurrent, unresectable, locally advanced or metastatic tumor lesions, and palliative care as the preferred treatment modality; (IV) tumor tissue sample obtained within 6 months of enrollment, or at least one tumor lesion accessible and permission granted for biopsy; (V) patients with multiple lines of prior therapy were eligible if their Eastern Cooperative Oncology Group (ECOG) performance status was 0–2; and (VI) voluntary participation in the study with a signed informed consent form.
Sample collection and targeted next-generation sequencing (tNGS)
For each participant who underwent tNGS, the submission of at least 20 to 40 unstained slides containing more than 20% tumor cells, a formalin-fixed paraffin-embedded (FFPE) block, or 0.5 cm3 fresh-frozen tissue was required, to extract at least 50 ng of DNA for each sample. These tumor tissues had to have been obtained within 6 months of the enrollment of the patient in the study. All samples were obtained as part of routine clinical care; the protocol did not mandate biopsies for research purposes. All tumor samples were reviewed by clinical pathologists who specialized in gastrointestinal tumors before tNGS.
Comprehensive genomic alteration analysis of the tumor and matched blood samples were performed with an assay panel that captured 450 cancer-related genes and selected introns of 38 genes frequently rearranged in cancer (YuansuTM, OrigiMed, listed in Table S1). With a mean coverage of 900× for tumor tissues and 300× for paired blood cells by an Illumina NextSeq-500 Platform (Illumina Incorporated, San Diego, CA).

Full table
Variations calling and annotation
Somatic alternations, including base substitutions, insertions and deletions, copy number alterations, and gene fusions/rearrangements, were identified as our previous report (11). The bioinformatic protocol is similar with previous published study (12). Briefly, reads were aligned to human genome with the reference sequence (hg19) by Burrows-Wheeler Aligner (BWA), accompanying with removing duplicates from polymerase chain reaction (PCR) by Picard. MuTect identified SNVs and short indels after quality recalibration and realignment using Genome Analysis Toolkit (GATK) pipeline. Pindel was used to calibrate short indels. Read depths were normalized in target regions by Exome Copy number Alterations/Variations annotATOR (EXCATOR). Customized algorithms were used to detect copy number changes and gene rearrangement. For copy-number variations (CNVs), genes with a threshold of surpassing 4-copies are deemed as amplification, and genes with 0-copies are homozygous deletions. Germline variants were identified by HaplotypeCaller from the Genome Analysis Toolkit (GATK v.3/3) in the gvcf mode with default settings (13), and only those present in both normal and tumor samples were retained.
For annotations of actionability for each alteration, several databases were refereed, including OncoKB knowledge database (14), DGIdb (v3.0.2) (15) and PanDrugs (version: 2018.11.7) (16). The functional significance of variants was also determined by interrogating databases and published literature, such as ClinVar, Catalogue of Somatic Mutations in Cancer (COSMIC), and PubMed.
MDTB review and management
Clinical information and genomic alterations results were reviewed by a MDTB that consisted of several professors specializing in general surgery, oncology, radiology, radiotherapy, pathology, genome bioinformatics and nursing care.
In the clinical management of patients with actionable alterations and targeted therapy, a recommendation was made if the matched targeted therapy was available via a clinical trial or as an approved drug with an appropriate dose and cycle. Actionable alterations were defined as any genomic mutation in a gene or a pathway of a gene that is directly targeted by an approved drug.
The patient needed to have been in adequate physical condition to receive treatment. For patients without actionable alterations or who were not in adequate condition to receive matched targeted therapy or for those who rejected actionable alteration-based treatment, the MDTB formulated a therapeutic regimen combined with best care support for each patient; these regimens included chemotherapy, multitarget molecular targeted drugs and regional treatment such as radiotherapy or ablation. The therapeutic regimen was evidence-based according to clinical treatment guidelines and experience, and the adoption of the regimen was determined by each patient.
Immunohistochemistry (IHC)
IHC was performed on FFPE specimens. Serial 4-µm-thick sections were sliced and placed on glass slides for IHC staining. The following primary antibodies were used: anti-CDKN2A p16INK4A (mouse monoclonal 5A8A4, dilution: 1/1,000, Invitrogen, US) and anti-Her-2 (mouse monoclonal ZM-0065, Zhong-shan Inc., China). Secondary antibodies were added to all the sections, including negative control slides, which omitted the primary antibody treatment.
The evaluation of protein expression was performed by independent pathologists who were blinded to the clinicopathologic data. The criteria for positivity were as follows. For Her-2, the evaluation standard was according to the College of American Pathologist/American Society of Clinical Oncology (CAP/ASCO) criteria for breast cancer (17). For CDKN2A, the samples were classified as the following two types: “retained” and “lost”. Samples with nuclear and/or cytoplasmic expression anywhere within the tumor were defined as “retained”, while the other samples were defined as “lost” if there was completed deficiency of protein expression in the tumor (18).
Clinical data collection and follow-up
Clinical and demographic data were collected at the time of initial enrollment, and these data included age, diagnosis, therapeutic history, current disease status, pathological status, laboratory examination results (full blood count, liver and renal function, coagulation and tumor biomarker CEA/CA19-9/AFP/CA125) and the latest imaging examinations. For patient follow-up, radiological evaluations were performed every 6–8 weeks, and the outcomes were analyzed using the RECIST1.1 (19).
Statistical analysis
Statistical analysis was performed using R version 3.1.1. Continuous variables and proportions were compared using the Mann-Whitney, chi-squared and Fisher’s exact tests, as appropriate. The 95% CIs were constructed using the customary normal approximation interval formula. Correlations between marker expression levels were analyzed by Spearman’s rank correlation coefficient. A two-tailed P<0.05 was considered statistically significant.
Results
Patient enrollment and characteristics
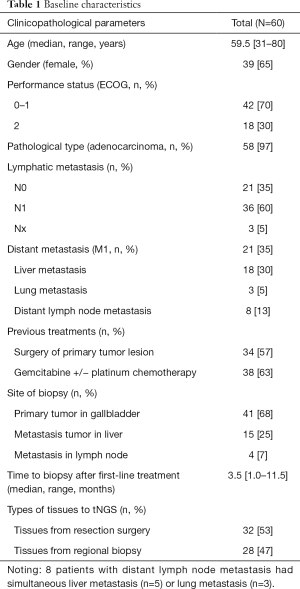
From March 2016 to March 2018, 60 patients with GBC applied for admission to our study. Among these participants, the median age was 59.5 (range: 31–80) years, with 39 females and 21 males. In total, 97% (58/60) of the patients were diagnosed with gallbladder adenocarcinoma, and 2 patients were diagnosed with mixed adeno-endocrine carcinoma (Table 1).
Full table
Tumor tissues obtained within six months of enrollment in the trial were used for ultra-deep tNGS. For patients without tumor tissues available from within six months of their enrollment in the trial, a biopsy of primary tumor lesions was needed to acquire recent tissue. Samples from all participants with pathologically confirmed GBC underwent tNGS, and the treatment and management of all 60 participants with GBC were managed by the MDTB. With regard to prior therapies, all patients undergone at least a line of antitumor treatments, 34 (57%) patients had undergone surgical resection, and 38 (63%) patients had received gemcitabine and/or platinum chemotherapy (Table 1). At the time of enrollment, 36 (60%) patients had evidence of lymphatic metastasis, and 21 (35%) patients had extra-gallbladder metastatic sites. To maximize the efficiency of tNGS and its application to the clinical management of GBC patients, the mean wait time for tNGS reports is 12 days (range 7 to 16 days), and MDTB recommendations are provided within 16 days on average (range 10 to 21 days), including the wait time for the tNGS reports.
Spectrum of genomic alterations in GBC
Using tNGS (mean coverage depth: 986×) to capture exons of cancer-related genes (n=450) and selected introns of 38 oncogenes or tumor suppressor genes, all 60 GBC patients were found to have at least one somatic nonsynonymous mutation. The spectra of somatic mutations and the tumor mutation burdens (TMBs) varied among samples, indicating heterogeneity among GBC patients. The median TMB was 5.4 (range: 0.8–36.74) mutations/Mb. The TMB found in our study was significantly higher than that found in patients from TCGA-CHOL with cholangiocarcinoma (20) and that found in patients with GBC evaluated by Li et al. (6,21); these differences may result from the use of samples from patients with late clinical stage and post-chemotherapy tumors in our GBC cohort. Consistent with the findings of previous studies (22), we identified some genes with high mutation frequencies in GBC, including known oncogenes (PI3KCA and KRAS) and tumor suppressor genes (TP53, CDKN2A, ARID1A, and APC). The most frequently altered genes among the 60 patients with GBC in this study were TP53 (73%), CDKN2A (25%), PIK3CA (20%) and ERBB2 (18%) (Figure 2). Interestingly, we identified one patient (PUHG001) who carried the POLEN2126S mutation and who had a TMB of 34.2 mut/Mb. The mutant IDH alleles detected in our cohort (IDH1R132L) were different from those found in TCGA-CHOL (IDH1R132C and IDH2R172K/S). Moreover, we found a high frequency of truncation mutations in SWI/SNF complex genes, such as ARID1A, ARID2 and PBRM1. A BAP1 truncation mutation was identified in 2 patients. We also identified 4 GBC patients with somatic amplification of mouse double minute 2 (MDM2), who were unsuitable candidates for immune checkpoint inhibitors because of the potential risk of hyper-progression (23).
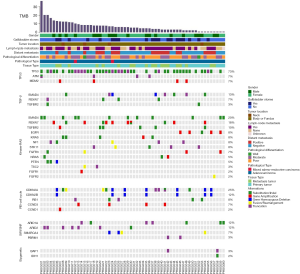
We identified the mutational features of each frequently mutated gene. TP53 and PIK3CA predominately contained oncogenic base substitutions and indels (insertion or deletion), CDKN2A and CDKN2B predominantly contained homozygous deletions, and ERBB2 mutations were mostly gene amplifications. CDKN2A had mostly deletions in the p16INK4 region (46.7%, 7/15), and CDKN2B had only one deletion mutation type in the p15INK4b region. Functional pathways of the somatic alterations (mutations and copy-number variations) revealed that the cell cycle, SWI/SNF complex and kinase functions in the Ras pathway were frequently altered in GBC patients (Figure 2).
IHC assessment and survival outcomes for CDKN2A and ERBB2
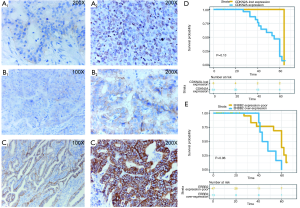
Prior outcomes indicated that GBC patients may be a suitable population for targeted-CDKN2A [palbociclib (24)] or anti-ERBB2 (Her-2) therapy (25). In our present cohort, 7 patients were identified as having deleterious mutations in CDKN2A (CDKN2ADel), while 8 patients had amplification of ERBB2 (ERBB2-amp). Because alterations in protein expression are also indicators for targeted therapy, it is also important to explore CDKN2A and ERBB2 expression in situ in tumor tissue by IHC (Figure 3A,B,C). Using FFPE samples from the present cohort, we evaluated CDKN2Ap16 and ERBB2 protein expression. CDKN2A expression was lost (Figure 3A, 1) in 10% (6/60, 6/7 patients with CDKN2A-del) of the patients with GBC, and 8.3% (5/60, 5/8 patients with ERBB2-amp) of the patients had ERBB2 overexpression (3+ by IHC, Table 2, Figure 3C, 1/2). This finding indicates that a small proportion of GBC patients may be suitable for palbociclib or anti-Her-2 targeted treatment (26). For survival analysis, we did not observe any prognostic significance of or survival correlation with CDKN2A (P=0.13) and ERBB2 (P=0.06) (Figure 3D,E).

Full table
Actionable fusion events and germline mutations in relation to family history of malignancies
We captured gene fusion events in select introns of 38 genes to identify virtual fusion events and fusion drivers (Figure 4). In total, 23% (14/60) of the patients had gene fusion events, with 17 fusion events captured. 14 of the 17 fusion events were accompanied by cancer-driving genes. FGFR-related fusion genes, which have been previously reported in biliary tract cancers (BTCs), were deemed to be the drivers of BTC development (27). In our GBC cohort, we identified FGFR-related fusions in 3% (2/60) of the patients; these fusions involved FGFR3-TACC3 and in-frame fusions of FGFR2. The FGFR3-TACC3 fusion was previously reported in cholangiocarcinoma (27) and was shown to be oncogenic in lung cancer, glioblastoma, and cervical cancer (28,29). To the best of our knowledge, this is the first report of this fusion in GBC. Interestingly, one NTRK3 fusion event, thought to be a rare event in cancer, was captured in patient PUHG054; that patient may benefit from larotrectinib treatment (a highly selective TRK inhibitor) (30).
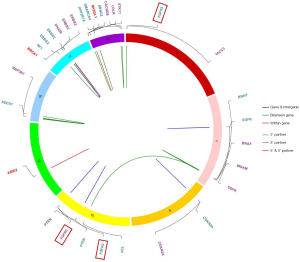
Germline pathogenic alterations were also identified in all patients who underwent tNGS. We analyzed and annotated the significance of germline variants in 10 DNA repair-associated genes, including ATM, BRCA1/2, FANCA, MLH1, MSH2, MSH6, PMS2, PALB2, and RAD5, and in 3 genes related to genomic instability (TP53, BAP1, BLM). In total, 2 of the 60 patients carried deleterious germline mutations. The genes with deleterious pathogenic germline mutations were ATM and BRCA2. Additionally, 25 germline mutations in 20 other GBC patients were identified as variants of uncertain significance (VUS). Only 6 of the 22 patients with germline alterations reported a family history of cancer, and none of the patients reported family members with GBC. Among the 6 patients with a family history of cancer, only 1 patient (with deleterious BRCA2 mutations) had a positive family history of cancer in first-degree relatives.
Clinical translation and practice
For each patient enrolled in this study, after identifying the individuals’ genomic alterations by tNGS, we integrated the genomic information and their clinicopathological features to explore the translational percentage of mutation-targeted therapy. To annotate the actionability for each identified alteration, we comprehensively considered the evidences and recommendations from three major databases, including OncoKB, DGIdb and PanDrugs (see Methods), and then we determined four levels of evidence for the altered genes identified in our cohort, according to existing clinical studies for each mutation and its corresponding targeted drugs (level 1 GPTT indicated the strongest recommendation while level 4 GPTT indicated the weakest recommendation, Table S2). Overall, 46 of the 60 (76%) patients were matched with at least one GPTT regimen, among whom 26 (43.3%) GBC patients were matched with two or more GPTT options. Although there were only one level-1 GPTT case with NTRK3 fusion event, 18.3% (11/60) of the patients possessed level 2 recommendations. Among these actionable genomic alterations, the most common actionable targets were PIK3CA, ERBB2/3 and CDKN2A, and the most common variation types were somatic missense mutations and copy number variations. Pathway-enrichment analysis demonstrated that the most common actionable molecular mechanisms were related to the PI3K-Akt signaling pathway. However, for the limitation of available molecular targeted drugs during the real-world clinical practice, only 2 patients proceeded GPTT regimen to receive therapy targeted the specific genomic alterations. PUHG006, a 54 years old woman with advanced gallbladder adenocarcinoma, was detected a CDKN2A fragment deletion in p16INK4A. After failed in the treatment of GEMOX chemotherapy and radiotherapy, she received palbociclib treatment for 4 weeks. The CA19-9 significantly decreased (from 1891 to 821) while she discontinued therapy because of adverse events (thrombocytopenia, grade 3; diarrhea, grade 3). Another previously reported patient (31), PUHG038, with germline BRCA1Q858* mutation, treated by olaparib and obtained 10-weeks tumor progression-free survival.
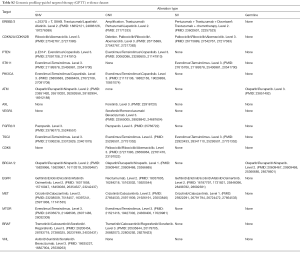
Full table
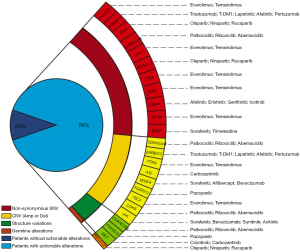
From the actionable genomic alterations revealed by tNGS, we constructed the GPTT landscape for precision oncology in GBC patients (Figure 5). We systematically reviewed the published literature and molecular targeted drugs listed on the market and established the framework for matching each genomic alteration with a GPTT regimen. This landscape will offer a reference for the clinical practice of and trial design for precision oncology within GBC.
Discussion
GBC is a rare cancer with poor prognosis, short survival time and limited therapeutic regimens. This rare tumor has not been well studied. Precision oncology becomes feasible when the genomic landscape of GBC becomes clearer. However, limited efforts have been made to explore the genomic alteration landscape of this rare tumor and to develop guidelines for the clinical practice of precision oncology in GBC (32). Therefore, we conducted a prospective proof-of-concept trial to assess the feasibility of using molecular targeted agents guided by tNGS in a real-world clinical practice. We identified at least one molecular aberration based on tNGS in each of the 60 enrolled patients with refractory, which is a promising outcome for clinical practice, as GBC has low tumor cellularity and high tumor stroma. Among these genomic alterations, we found that 76% of the patients carried at least one actionable alteration that could be matched with molecular targeted drugs listed on the market, potentially enabling the application of precision targeted therapy (Figures 2,5). The mapping rate from the tNGS results to the targeted therapy regimens was higher than the rates observed in the SHIVA (33) trial (39%) and MOSCATO-01 (34) trial (24%), while the MOSCATO-01 trial concluded that 68% of the patients with bile tract cancers (33/43) could match targeted therapy through tNGS (35). These outcomes demonstrated that GBC was a particularly well-suited candidate for precision medicine programs based on molecular or genomic alterations (4). To the best of our knowledge, the 60 GBC patients included in this study was a relatively large sample size for this rare tumor.
The clinical benefit of GPTT in GBC patients is worth investigating. Loic Verlingue et al. showed encouraging results of the application of the GPTT regimen in the MOSCATO-01 trial, with a 33% overall response rate and 88% DCR for 18 patients with advanced biliary tract cancers and a median PFS of 5.2 months (35). In addition, in the present study, we also validated the clinical significance of alterations in CDKN2A and ERBB2 (or Her-2) detected by tNGS. In total, 11.7% of the GBC patients in this trial had CDKN2A deletions, and 10% had lost protein expression, as assessed by IHC; these patients were candidates for treatment with palbociclib. 8.3% patients had ERBB2 amplifications and were candidates for anti-Her2 therapy, including treatment with trastuzumab or lapatinib (25). Moreover, the promising efficacy of tumor immunotherapy in several solid tumors, especially that of immune-checkpoint inhibitors such as nivolumab and pembrolizumab, linked genomic alterations and clinical treatment outcomes more firmly (36). The TMB, a factor strongly correlated with PD-1 inhibitors, has been widely assessed before the initiation of anti-PD1/L1 therapy. In our previous clinical trial (37), we demonstrated that a TMB of 12.5 mutations/Mb could serve as the cutoff value for determining the therapeutic benefit of a regimen of lenvatinib plus PD-1 inhibitor. In the present study, we found that 15% (9/60) of patients could be classified as a TMB-high GBC, and this proportion of patients may benefit from immune-checkpoint inhibitor.
However, it is important to note that a high mapping rate does not indicate a high translation rate to practical clinical treatment. In our cohort, the enrolled patients had all previously received at least one treatment, with 63% of the participants experiencing disease progression after gemcitabine and/or platinum chemotherapy. Although 46 of the 60 patients had actionable genomic alterations and were matched to suitable molecular targeted agents, only 20% (12/60) could match a therapeutic regimen with level-1/2 GPTT. The limitation of our study was that only 2 patients accepted the GPTT regimen. Based on our experience, the obstacles to GPTT include the following: (I) the shortage of evidences of clinical practices using targeted drug based on actionable alterations in patients with rare tumors, such as everolimus in GBC patients with mutations in PI3K-Akt signaling pathway. (II) The poor physical conditions of advanced GBC patients that render them unable to tolerate anti-cancer treatment, especially patients with jaundice or severe liver dysfunction. (III) The high degree of heterogeneity of the GBC genome made it hard to specifically select targets to inhibit; thus, multitarget drugs may be more valuable when precise molecular agents targeting specific mutations (such as KRAS and TP53) are lacking.
Therefore, importantly, a low translation rate to clinical treatment requires assistance from a MDTB. The following three aspects reflect the value of a MDTB in GPTT: (I) for decision making in a GPTT-suitable population, the MDTB can recommend a therapy with reference to the existing evidence regarding genomic alterations and available clinical trial results, and it can weigh the options when different actionable targets match various molecular drugs; (II) for patients unable to accept GPTT, the MDTB offers recommendations for the optimal alternate therapeutic methods, highlighting the need for personalized treatment; and (III) best support care (BSC) is indispensable for patients with advanced bile tract cancers to improve their quality of life and support good physical condition to enable them to receive anti-tumor treatment.
In conclusion, in the present study, we identified the mutational spectrum and actionable targets in GBC, a rare cancer. With the assistance of the MDTB, GPTT guided by ultra-deep tNGS is feasible in real-world clinical practice, and the majority of GBC patients had actionable genomic alterations. The translation rate of GPTT in clinical treatment has substantial room for improvement, and larger-scale prospective clinical trials with interventional GPTT treatment are necessary to demonstrate the superiority and efficacy of GPTT among patients with refractory GBC.
Acknowledgments
We thank American Journal Experts Co., Ltd. in improving English writing and modifying grammar.
Funding: This work was supported by the International Science and Technology Cooperation Projects (2016YFE0107100 and 2015DFA30650), the CAMS Innovation Fund for Medical Science (CIFMS) (2017-I2M-4-003), the Beijing Natural Science Foundation (L172055), the National Ten-thousand Talent Program, the Beijing Science and Technology Cooperation Special Award Subsidy Project and the CAMS Initiative for Innovative Medicine (CAMS-2018-I2M-3-001).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethics Statement: The authors are accountable for all aspects of the work (if applied, including full data access, integrity of the data and the accuracy of the data analysis) in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patients participated in this observational trial provided written informed consent for medical treatment and collection of tumor biopsies and for comprehensive molecular profiling of tumor and germline samples. All patients were enrolled in institutional protocols approved by the Ethics Review Committee of Peking Union Medical College Hospital (Beijing, China) (reference No. PM-1503).
References
- Hundal R, Shaffer EA. Gallbladder cancer: epidemiology and outcome. Clin Epidemiol 2014;6:99-109. [PubMed]
- Rakić M, Patrlj L, Kopljar M, et al. Gallbladder cancer. Hepatobiliary Surg Nutr 2014;3:221-6. [PubMed]
- Valle J, Wasan H, Palmer DH, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 2010;362:1273-81. [Crossref] [PubMed]
- Valle JW, Lamarca A, Goyal L, et al. New Horizons for Precision Medicine in Biliary Tract Cancers. Cancer Discov 2017;7:943-62. [Crossref] [PubMed]
- Belin L, Kamal M, Mauborgne C, et al. Randomized phase II trial comparing molecularly targeted therapy based on tumor molecular profiling versus conventional therapy in patients with refractory cancer: cross-over analysis from the SHIVA trial. Ann Oncol 2017;28:590-6. [PubMed]
- Li M, Liu F, Zhang F, et al. Genomic ERBB2/ERBB3 mutations promote PD-L1-mediated immune escape in gallbladder cancer: a whole-exome sequencing analysis. Gut 2019;68:1024-33. [Crossref] [PubMed]
- Wardell CP, Fujita M, Yamada T, et al. Genomic Characterization of Biliary Tract Cancers Identifies Driver Genes and Predisposing Mutations. J Hepatol 2018;68:959-69. [Crossref] [PubMed]
- Cai W, Yuan Y, Ge W, et al. EGFR Target Therapy Combined with Gemox for Advanced Biliary Tract Cancers: a Meta-analysis based on RCTs. J Cancer 2018;9:1476-85. [Crossref] [PubMed]
- Saha SK, Gordan JD, Kleinstiver BP, et al. Isocitrate Dehydrogenase Mutations Confer Dasatinib Hypersensitivity and SRC Dependence in Intrahepatic Cholangiocarcinoma. Cancer Discov 2016;6:727-39. [Crossref] [PubMed]
- Javle M, Lowery M, Shroff RT, et al. Phase II Study of BGJ398 in Patients With FGFR-Altered Advanced Cholangiocarcinoma. J Clin Oncol 2018;36:276-82. [Crossref] [PubMed]
- Lin J, Shi J, Guo H, et al. Alterations in DNA Damage Repair Genes in Primary Liver Cancer. Clin Cancer Res 2019;25:4701-11. [Crossref] [PubMed]
- Wen S, Dai L, Wang L, et al. Genomic Signature of Driver Genes Identified by Target Next-Generation Sequencing in Chinese Non-Small Cell Lung Cancer. Oncologist 2019. [Epub ahead of print]. [Crossref] [PubMed]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491-8. [Crossref] [PubMed]
- Chakravarty D, Gao J, Phillips SM, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017;2017.
- Cotto KC, Wagner AH, Feng YY, et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res 2018;46:D1068-73. [Crossref] [PubMed]
- Piñeiro-Yáñez E, Reboiro-Jato M, Gomez-Lopez G, et al. PanDrugs: a novel method to prioritize anticancer drug treatments according to individual genomic data. Genome Med 2018;10:41. [Crossref] [PubMed]
- Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013;31:3997-4013. [Crossref] [PubMed]
- Qian ZR, Rubinson DA, Nowak JA, et al. Association of Alterations in Main Driver Genes With Outcomes of Patients With Resected Pancreatic Ductal Adenocarcinoma. JAMA Oncol 2018;4:e173420. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). European Journal of Cancer 2009;45:228-47. [Crossref] [PubMed]
- Farshidfar F, Zheng S, Gingras MC, et al. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles. Cell Rep 2017;18:2780-94. [Crossref] [PubMed]
- Li M, Zhang Z, Li X, et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat Genet 2014;46:872-6. [Crossref] [PubMed]
- Nakamura H, Arai Y, Totoki Y, et al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47:1003-10. [Crossref] [PubMed]
- Kato S, Goodman A, Walavalkar V, et al. Hyperprogressors after Immunotherapy: Analysis of Genomic Alterations Associated with Accelerated Growth Rate. Clin Cancer Res 2017;23:4242-50. [Crossref] [PubMed]
- Gao J, Adams RP, Swain SM. Does CDKN2A loss predict palbociclib benefit? Curr Oncol 2015;22:e498-501. [Crossref] [PubMed]
- Javle M, Churi C, Kang HC, et al. HER2/neu-directed therapy for biliary tract cancer. J Hematol Oncol 2015;8:58. [Crossref] [PubMed]
- Fujimura T, Takayama K, Takahashi S, et al. Estrogen and Androgen Blockade for Advanced Prostate Cancer in the Era of Precision Medicine. Cancers (Basel) 2018.10. [PubMed]
- Jusakul A, Cutcutache I, Yong CH, et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov 2017;7:1116-35. [Crossref] [PubMed]
- Tamura R, Yoshihara K, Saito T, et al. Novel therapeutic strategy for cervical cancer harboring FGFR3-TACC3 fusions. Oncogenesis 2018;7:4. [Crossref] [PubMed]
- Bielle F, Di Stefano AL, Meyronet D, et al. Diffuse gliomas with FGFR3-TACC3 fusion have characteristic histopathological and molecular features. Brain Pathol 2018;28:674-83. [Crossref] [PubMed]
- Drilon A, Laetsch TW, Kummar S, et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med 2018;378:731-9. [Crossref] [PubMed]
- Xie Y, Jiang Y, Yang XB, et al. Response of BRCA1-mutated gallbladder cancer to olaparib: A case report. World J Gastroenterol 2016;22:10254-9. [Crossref] [PubMed]
- Takeda H, Nishikawa H, Osaki Y. The new era of precision medicine in hepatocellular carcinoma: the urgent need for promising biomarkers. Hepatobiliary Surg Nutr 2018;7:490-1. [Crossref] [PubMed]
- Le Tourneau C, Delord JP, Goncalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 2015;16:1324-34. [Crossref] [PubMed]
- Massard C, Michiels S, Ferte C, et al. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov 2017;7:586-95. [Crossref] [PubMed]
- Verlingue L, Malka D, Allorant A, et al. Precision medicine for patients with advanced biliary tract cancers: An effective strategy within the prospective MOSCATO-01 trial. Eur J Cancer 2017;87:122-30. [Crossref] [PubMed]
- Miao D, Margolis CA, Vokes NI, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet 2018;50:1271-81. [Crossref] [PubMed]
- Lin J, Shi W, Zhao S, et al. Lenvatinib plus checkpoint inhibitors in patients (pts) with advanced intrahepatic cholangiocarcinoma (ICC): Preliminary data and correlation with next-generation sequencing. J Clin Oncol 2018;36:abstr 500.



