
Plasma miR-146a predicts serological conversion of hepatitis B e-antigen (HBeAg) in chronic hepatitis B patients treated with nucleotide analogs
Introduction
An estimated 240 million individuals worldwide have chronic hepatitis B (CHB) virus (HBV) infection, which results in approximately 780,000 annual deaths (1,2). Patients with CHB are at elevated risk of severe liver inflammation and fibrosis, which can progress to serious complications, such as cirrhosis, hepatic decompensation, and hepatocellular carcinoma (HCC) (3). The natural course of HBV infection depends on the presence of hepatitis B e-antigen (HBeAg), which is indicative of HBV replication (4,5). CHB patients with HBeAg (HBeAg-positive patients) are highly infectious and at high risk of developing liver cirrhosis, HCC, and other liver complications (6-9), and therefore treatment is recommended for such patients. HBeAg is thus a useful parameter for monitoring treatment and seroconversion of CHB, as well as the efficacy of antiviral therapies (10). HBeAg seroconversion is associated with disease remission, lower incidence of both liver cirrhosis and HCC, and higher survival rates (11-13). Nucleotide analogs (NAs) are antiviral drugs that block reverse transcriptase and thereby prevent viral replication. Telbivudine (LdT), an oral thymidine analog (L-nucleoside) with minimal hepatic metabolism, is primarily eliminated by renal clearance (14). While LdT yields a higher HBeAg seroconversion rate than other NAs (15-18), 20% of patients tend to develop LdT resistance within 2 years (19). Entecavir (ETV) is an oral guanosine analog used against HBV and one of the favorite first-line NAs because of its high barrier to resistance, which confers high probability of successful long-term therapy (20). LdT and ETV are among the 3 most potent drugs used against HBV (20). Adefovir dipivoxil (ADV) is an adenosine analog less potent than LdT and ETV; despite 29% of patients developing resistance at 5 years (20,21), it is nevertheless useful against lamivudine-resistant HBV (22) but associated with renal toxicity (23).
MicroRNAs are often involved in HBV replication, CHB-induced fibrosis, inflammatory liver diseases, and HCC (24-26). They are also used as predictive markers of HCC prognosis and response to therapy (27,28). MiR-146a plays a vital role in the control of T helper type 1 (Th1) response mediated by regulatory T cells (Tregs) by targeting signal transducer and activator transcription 1 (STAT1) (29), and it is involved in negative regulation of the toll-like receptor (TLR)-mediated inflammatory response (30). In addition, miR-146a participates in the modulation of T cell activation (31) and dendritic-cell homeostasis and functions (32,33) and also contributes to CHB-related physiological and pathological processes (34,35).
The rs2910164 single-nucleotide polymorphism of pre-miR-146a is negatively correlated with acute-on-chronic liver failure (ACLF)-HBV susceptibility in Chinese patients (36). Meanwhile, miR-146a expression is reportedly downregulated during the immune-tolerant (IT) phase of CHB, which might be related to immune tolerance (37). Furthermore, researchers have found a positive association between miR-146a and (alanine) transaminase ALT levels in patients during the immune-active (IA) phase of CHB, and miR-146a is notably upregulated in HBV-expressing HuH-7 hepatocytes, HBV-expressing mice, and patients with HBV infection (38). It has also been demonstrated that the HBx-miR-146a-complement factor H (CFH) activation regulation pathway might play an important role in the immunopathogenesis of CHB (38).
Recent data show that miR-146a can promote viral replication (39); nevertheless, its role in predicting CHB prognosis after antiviral therapy remains unclear. The present study therefore aimed to investigate the association of plasma miR-146a with serological conversion of HBeAg in CHB patients treated with NAs.
Methods
Study design and patients
This was a retrospective study of HBeAg-positive patients with CHB treated at Xiangya Hospital, Central South University, Changsha, China between September 2009 and March 2014. Inclusion criteria were: (I) hepatitis B surface antigen (HBsAg) and HBeAg positivity for >6 months; (II) baseline HBV deoxyribonucleic acid (DNA) levels ≥106 IU/mL; (III) ALT levels at least 2-fold above the upper limit of normal (ULN; 40 IU/L); and (IV) completion of the 96-week follow-up. Exclusion criteria were: (I) other hepatotropic viral infections; (II) alcoholic or non-alcoholic liver disease; (III) drug-induced hepatitis; (IV) autoimmune liver disease; or (V) cirrhosis.
This study was approved by the Medical Ethics Committee of Xiangya Hospital. The committee waived the requirement of individual consent due to the retrospective nature of the study. We obtained all of the blood samples from our institution’s biological specimen bank. All of the patients provided written informed consent for their samples to be archived in this biobank.
Grouping
The treatment course of NAs was 104 weeks. We measured serum ALT, HBV DNA, HBsAg, HBeAg, and anti-HBe before treatment and at 24, 48, 72, and 104 weeks after treatment began. We measured plasma miR-146a at 0, 24, 48, and 104 weeks of treatment, after which we calculated and analyzed ΔmiR-146a24w and ΔmiR-146a48w. Patients were grouped according to whether they had achieved HBeAg seroconversion by the 104th week of treatment. HBeAg seroconversion was defined as HBeAg clearance and absence of anti-HBe antibodies in previously HBeAg-positive CHB patients.
Laboratory indexes
We measured ALT on an Olympus AU640 Automatic Biochemical Analyzer (Olympus, Tokyo, Japan). The reference range for ALT was 0–40 U/L. We detected serum HBsAg, HBeAg, and anti-HBe by commercial chemiluminescent microparticle immunoassays [CIMA; catalog nos. HBsAg 31587LIF00, HBeAg 31483, hepatitis B e-antibody (HBeAb) 29560LZ01; Abbott Laboratories, Abbott Park, Illinois, USA] undertaken on an i2000 system (Abbott Labs). Positivity was defined as >0.05 IU/mL for HBsAg, >1.0 s/co for HBeAg, and <1.0 s/co for HBeAb. HBV DNA was measured with a real-time (RT) fluorescence quantitative polymerase chain reaction (PCR) kit (Roche Diagnostics, Basel, Switzerland) on a 7500 RT quantitative thermocycler (Applied Biosystems, Foster City, California, USA). The lower limit of detection was 69.84 IU/mL (40).
Real-time quantitative PCR (qRT-PCR)
We collected whole blood (10 mL) from fasting patients in heparin sodium anticoagulation tubes and then centrifuged the blood samples at 3,000 rpm for 15 min to obtain plasma. We extracted total RNA from plasma with the miRNeasy Serum/Plasma Kit (Qiagen, Venlo, The Netherlands) per manufacturer’s instructions. Only RNA samples with an OD260/OD280 ratio of 1.8–2.0 were used for RT-PCR.
Next, we synthesized complementary DNA (cDNA) from total RNA on a miScript PCR system (QuantiTect Reverse Transcription Kit, Qiagen Co., Germany). The reaction mixture (20 µL) contained 1.5 µL RNA, 4 µL 5× miScript HiSpec Buffer, 2 µL 10× miScript Nucleics Mix, 10.5 µL RNase-free water, and 1 µL miScript Reverse Transcriptase mix. We performed amplification at 37 °C for 60 min and 95 °C for 5 min. We used RNase-free water instead of RNA for negative control. Next, we used the obtained cDNA as a template for RT-PCR. As general-reference miRNAs such as U6 are not stable in plasma, we used cel-miRNA-39 derived from C. elegans (41) as an exogenous qRT-PCR primer. RT-PCR was performed with the miScript SYBR Green PCR Kit (Qiagen) per manufacturer’s instructions. All of the experiments were performed in triplicate. The amplification protocol was: (I) 95 °C for 15 min; and (II) 45 cycles of 94 °C for 15 s, 55 °C for 30 s, and 70 °C for 30 s. We conducted qRT-PCR on a StepOnePlus PCR Thermocycler (Life Technologies Co., Grand Island, New York, USA) and assessed miRNA expression by the 2−ΔΔCtmethod. The miR-146a primer was 5'-CCUCUGAAAUUCAGUUCUUCAG-3' (Qiagen). The cel-miRNA-39 primer was 5'-UCACCGGGUGUAAAUCAGCUUG-3' (Qiagen). We measured miR-146a at 24 and 48 weeks, but not at 104 weeks due to unavailability of blood samples. Changes in miR-146a were determined as follows:
- ΔmiR-146a48w = (miR-146a at 48 weeks − miR-146a baseline)/miR-146a baseline;
- ΔmiR-146a24w = (miR-146a at 24 weeks − miR-146a baseline)/miR-146a baseline.
Statistical analysis
An independent biostatistician analyzed all of the data using SPSS software version 18.0 (IBM, Armonk, New York, USA). Continuous data are mean ± standard deviation and were analyzed by Student’s t-test. Categorical data are presented as frequency and were analyzed by chi-square test. Patients were categorized based on presence/absence of HBeAg seroconversion at 104 weeks. We performed univariable and multivariable logistic-regression analyses to analyze and identify independent prognostic factors. We plotted receiver operating characteristic (ROC) curves to assess the prognostic value of miR-146a at 24 and 48 weeks. Two-sided P<0.05 was considered to be statistically significant.
Results
Baseline characteristics of patients
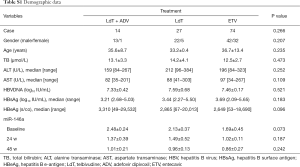
Of included patients, 41 were treated with 600 mg/day LdT (Novartis, Basel, Switzerland), and 74 were administered 0.5 mg/day ETV (ChiataiTianqing Co., Jiangsu, China). Fourteen patients treated with LdT had HBV DNA levels >300 IU/mL at 24 weeks and were therefore administered ADV (Chiatai Tianqing) based on response-guided therapy (RGT). Patient baseline characteristics were comparable between both groups, including gender, age, total bilirubin, ALT, AST, HBV DNA, HBsAg, HBeAg, and treatment response (all P>0.05; Table 1).
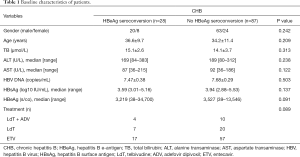
Full table
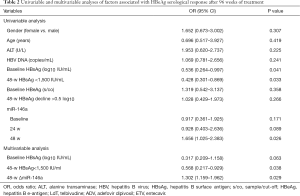
Expression of miR-146a
We measured miR-146a expression before treatment and after 24 and 48 weeks of treatment. As shown in Figure 1, miR-146a expression was significantly lower in the HBeAg seroconversion group after 24 and 48 weeks of treatment, compared with the HBeAg non-seroconversion group (P<0.01). Patients with no HBeAg serological conversion (87/115, 75.65%) was 3.1 folds the patients with HBeAg serological conversion (24.35%).
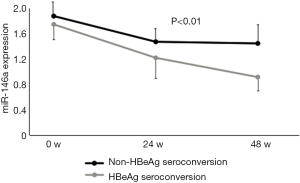
Univariable and multivariable analyses of factors associated with seroconversion of HBeAg after 104 weeks of treatment
In univariable analysis, HBsAg [odds ratio (OR) =0.536; 95% confidence interval (CI), 0.264–0.997; P=0.041], ΔmiR-146a48w (OR =1.656; 95% CI, 1.025–2.383; P=0.026), and 48w HBsAg <1,500 IU/mL (OR =0.428; 95% CI, 0.301–0.869; P=0.033) were associated with HBeAg seroconversion at 104 weeks (Table 2). In a multivariable model including all 3 variables, ΔmiR-146a48w (OR =1.302; 95% CI, 1.159–1.962; P=0.029) and 48w HBsAg <1,500 IU/mL (OR =0.568; 95% CI, 0.217–0.929; P=0.038) were independently associated with increased odds of HBeAg seroconversion at 104 weeks (Table 2).
Full table
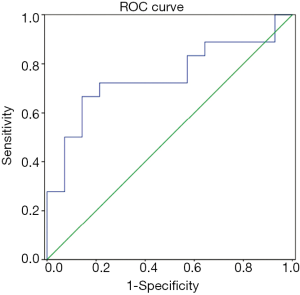
ROC curve of ΔmiR-14648w for HBeAg seroconversion
The area under the ROC curve (AUC) for ΔmiR-14648w was 0.757 for seroconversion of HBeAg (P=0.013; Figure 2). At the optimal cutoff value equivalent to a Youden index of 67.9%, the specificity and sensitivity of ΔmiR-14648w were 63.7% and 88.3%, respectively. Positive (PPV) and negative (NPV) predictive values were 70.87% and 84.48%, respectively.
Subgroup analysis
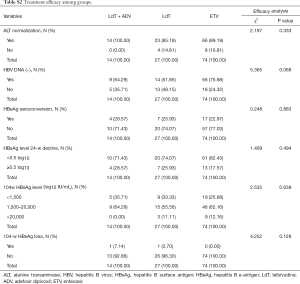
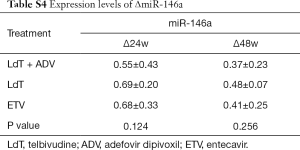
We next evaluated whether miR-14648w expression varied among patients treated with LdT, ETV, and LdT + ADV. As shown in Table S1, demographic data of patients undergoing different treatments were comparable (all P>0.05). There were no differences in efficacy among the different antiviral drugs (all P>0.05; Table S2). In addition, the effects of the antiviral drugs on miR-146a and ΔmiR-146a were comparable (all P>0.05; Tables S3,S4).
Full table
Full table

Full table
Full table
These results suggested that miR-146a was unaffected by NAs. Accordingly, miR-146a might be a biological marker for HBeAg seroconversion in HBeAg-positive patients treated with NAs.
Discussion
The present study showed that ΔmiR-146a48w was independently associated with seroconversion of HBeAg in CHB patients treated with NAs for 104 weeks. Moreover, ΔmiR-146a48w was universal, regardless of the type of NA administered. Taken together, these findings indicated that ΔmiR-146a48w could be a viable prognostic marker of seroconversion in CHB patients treated with NAs.
Previous studies have shown that miR-146a is upregulated in patients with HBV infection (38) and positively associated with ALT levels in patients during the IA phase of CHB (37). MiR-146a is therefore considered a potential circular marker of HBV infection. Our univariable and multivariable analyses suggested that ΔmiR-14648w could predict seroconversion of HBeAg after treatment in HBeAg-positive patients with CHB. To the best of our knowledge, this is the first study reporting decreased plasma miR-146a levels during LdT treatment for CHB. Notably, baseline miR-146a levels were not associated with HBeAg seroconversion, unlike ΔmiR-146a48w levels. Nor did we find any differences among the treatment regimens in ALT, HBV DNA, HBeAg seroconversion, or HBsAg, indicating that all 3 strategies were similarly successful for the management of this specific group of patients (42). Furthermore, we observed no differences in miR-146a and ΔmiR-146a48w among patients administered LdT, ETV, and LdT + ADV. These are therefore probable disease markers. Nevertheless, additional studies are needed to examine this relationship in a larger cohort of patients, including cases who develop resistance to NAs and those who fail treatment. HBV infection triggers a prolonged immune response involving innate immunity (43). In the virus’s pathogenesis, miR-146a could play a dominant immune-modulatory role.
Previous studies have revealed a positive correlation between miR-146a and ALT (37,44). However, miR-146a is reportedly not abnormally expressed in HBV carrier (45). In addition, it was shown that a panel of 11 miRNAs (not including miR-146a) is predictive of a sustained response to interferon treatment (46). Several reasons could explain these discrepancies. First, many cytokines, including tumor necrosis factor alpha (TNF-α) and interleukin-1 beta (IL-1β), are generally elevated in patients with HBV infection and might alter the expression of miR-146a (30,47). In the present study, miR-146a downregulation resulted from LdT treatment, which could also affect the expression of cytokines and other proteins. Second, different studies have taken different approaches to miRNA measurement and patient treatment. Therefore, miR-146a is probably a key molecule with the potential to influence HBV infection, but further comprehensive studies are required to assess the roles it plays in HBV-related innate immunity and tumorigenesis.
The present study had several limitations. First, the results may be influenced by biases inherent to retrospective studies (e.g., selection bias). Although we could not rule out potential selection bias, it likely had no major effect on the association between early changes in miR-146a24w and seroconversion of HBeAg. Second, despite our efforts to enroll as many patients as possible, the prolonged follow-up of 104 weeks resulted in a small sample size. A larger number of patients would provide higher statistical power. Third, although previous studies have shown that various HBV genotypes have very important effects on response to antiviral therapy (48-50), we were unable to determine HBV genotype due to the small sample size. Further prospective, long-term observational studies with larger sample sizes are needed to address these issues.
In summary, ΔmiR-146a48w was independently associated with HBeAg seroconversion in CHB patients who were HBeAg-positive after 104 weeks of treatment with NAs. ΔmiR-146a48w could be a prognosis marker of seroconversion in such patients.
Acknowledgments
Funding: This work was supported by grants from the 11th Five-Year National Science and Technology Major Projects (grant number: 2008ZX10002004), the 12th Five-Year National Science and Technology Major Projects (grant number: 2012ZX10002003). Hunan Natural Science Foundation (grant number: 2018JJ2661).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Medical Ethics Committee at the Xiangya Hospital of Central South University (No. 201408081, 201508104). The need for individual consent was waived by the committee owing to the retrospective nature of the study. All blood samples were from our biological specimen bank. All patients provided written informed consent for their samples to be archived in this biobank.
References
- Ott JJ, Stevens GA, Groeger J, et al. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine 2012;30:2212-9. [Crossref] [PubMed]
- Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012;380:2095-128. [Crossref] [PubMed]
- Koumbi L. Current and future antiviral drug therapies of hepatitis B chronic infection. World J Hepatol 2015;7:1030-40. [Crossref] [PubMed]
- Alexander W. 47th European Association for the Study of the Liver (EASL)/The International Liver Conference. P T 2012;37:362-3. [PubMed]
- Hadziyannis SJ, Papatheodoridis GV. Hepatitis B e antigen-negative chronic hepatitis B: natural history and treatment. Semin Liver Dis 2006;26:130-41. [Crossref] [PubMed]
- Papatheodoridis GV, Manolakopoulos S, Dusheiko G, et al. Therapeutic strategies in the management of patients with chronic hepatitis B virus infection. Lancet Infect Dis 2008;8:167-78. [Crossref] [PubMed]
- Papatheodoridis GV, Manolakopoulos S, Archimandritis AJ. Current treatment indications and strategies in chronic hepatitis B virus infection. World J Gastroenterol 2008;14:6902-10. [Crossref] [PubMed]
- Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology 2009;50:661-2. [Crossref] [PubMed]
- Liu D, Cui L, Wang Y, et al. Hepatitis B e antigen and its precursors promote the progress of hepatocellular carcinoma by interacting with NUMB and decreasing p53 activity. Hepatology 2016;64:390-404. [Crossref] [PubMed]
- Zhang X, Lin SM, Ye F, et al. An early decrease in serum HBeAg titre is a strong predictor of virological response to entecavir in HBeAg-positive patients. J Viral Hepat 2011;18:e184-90. [Crossref] [PubMed]
- Guidelines for the Prevention, Care and Treatment of Persons with Chronic Hepatitis B Infection. WHO Guidelines Approved by the Guidelines Review Committee. Geneva, 2015.
- Hsu YS, Chien RN, Yeh CT, et al. Long-term outcome after spontaneous HBeAg seroconversion in patients with chronic hepatitis B. Hepatology 2002;35:1522-7. [Crossref] [PubMed]
- Liaw YF. HBeAg seroconversion as an important end point in the treatment of chronic hepatitis B. Hepatol Int 2009;3:425-33. [Crossref] [PubMed]
- Zhou XJ, Swan S, Smith WB, et al. Pharmacokinetics of telbivudine in subjects with various degrees of renal impairment. Antimicrob Agents Chemother 2007;51:4231-5. [Crossref] [PubMed]
- Ma L, Cai YJ, Yu L, et al. Treatment with telbivudine positively regulates antiviral immune profiles in Chinese patients with chronic hepatitis B. Antimicrob Agents Chemother 2013;57:1304-11. [Crossref] [PubMed]
- Zhang Y, Lian JQ, Li Y, et al. Telbivudine plus adefovir therapy for chronic hepatitis B patients with virological breakthrough or genotypic resistance to telbivudine. Eur J Gastroenterol Hepatol 2013;25:814-9. [Crossref] [PubMed]
- Yan Z, Zhou J, Zhang M, et al. Telbivudine decreases proportion of peripheral blood CD4+CD25+CD127low T cells in parallel with inhibiting hepatitis B virus DNA. Mol Med Rep 2014;9:2024-30. [Crossref] [PubMed]
- Zheng Y, Huang Z, Chen X, et al. Effects of telbivudine treatment on the circulating CD4(+) T-cell subpopulations in chronic hepatitis B patients. Mediators Inflamm 2012;2012:789859. [Crossref] [PubMed]
- Liaw YF, Gane E, Leung N, et al. 2-Year GLOBE trial results: telbivudine Is superior to lamivudine in patients with chronic hepatitis B. Gastroenterology 2009;136:486-95. [Crossref] [PubMed]
- Menéndez-Arias L, Álvarez M, Pacheco B. Nucleoside/nucleotide analog inhibitors of hepatitis B virus polymerase: mechanism of action and resistance. Curr Opin Virol 2014;8:1-9. [Crossref] [PubMed]
- Chang TT, Gish RG, de Man R, et al. A comparison of entecavir and lamivudine for HBeAg-positive chronic hepatitis B. N Engl J Med 2006;354:1001-10. [Crossref] [PubMed]
- Lampertico P, Vigano M, Manenti E, et al. Adefovir rapidly suppresses hepatitis B in HBeAg-negative patients developing genotypic resistance to lamivudine. Hepatology 2005;42:1414-9. [Crossref] [PubMed]
- Fung J, Lai CL, Seto WK, et al. Nucleoside/nucleotide analogues in the treatment of chronic hepatitis B. J Antimicrob Chemother 2011;66:2715-25. [Crossref] [PubMed]
- Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol 2013;10:542-52. [Crossref] [PubMed]
- Jin BX, Zhang YH, Jin WJ, et al. MicroRNA panels as disease biomarkers distinguishing hepatitis B virus infection caused hepatitis and liver cirrhosis. Sci Rep 2015;5:15026. [Crossref] [PubMed]
- Wen Y, Peng SF, Fu L, et al. Serum levels of miRNA in patients with hepatitis B virus-associated acute-on-chronic liver failure. Hepatobiliary Pancreat Dis Int 2018;17:126-32. [Crossref] [PubMed]
- Wong CM, Wong CC, Lee JM, et al. Sequential alterations of microRNA expression in hepatocellular carcinoma development and venous metastasis. Hepatology 2012;55:1453-61. [Crossref] [PubMed]
- Boutz DR, Collins PJ, Suresh U, et al. Two-tiered approach identifies a network of cancer and liver disease-related genes regulated by miR-122. J Biol Chem 2011;286:18066-78. [Crossref] [PubMed]
- Lu LF, Boldin MP, Chaudhry A, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 2010;142:914-29. [Crossref] [PubMed]
- Taganov KD, Boldin MP, Chang KJ, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 2006;103:12481-6. [Crossref] [PubMed]
- Curtale G, Citarella F, Carissimi C, et al. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood 2010;115:265-73. [Crossref] [PubMed]
- Turner ML, Schnorfeil FM, Brocker T. MicroRNAs regulate dendritic cell differentiation and function. J Immunol 2011;187:3911-7. [Crossref] [PubMed]
- Ghani S, Riemke P, Schonheit J, et al. Macrophage development from HSCs requires PU.1-coordinated microRNA expression. Blood 2011;118:2275-84. [Crossref] [PubMed]
- Chen L. Expression and function analysis of microRNAs in peripheral blood mononuclear cells of patients with chronic hepatitis B virus infection. Central South University, 2012.
- Ge SF, Liu F, Xie JP. Advances in research on the mechanism of miRNAs regulating biological activity of hepatic stellate cells. Shandong Medical Journal 2016;56:101-3.
- Jiang H, He X, Li J, et al. Association of a single-nucleotide polymorphism within the miR-146a gene with susceptibility for acute-on-chronic hepatitis B liver failure. Immunogenetics 2013;65:257-63. [Crossref] [PubMed]
- Xing T, Xu H, Yu W, et al. Expression profile and clinical significance of miRNAs at different stages of chronic hepatitis B virus infection. Int J Clin Exp Med 2015;8:5611-20. [PubMed]
- Li JF, Dai XP, Zhang W, et al. Upregulation of microRNA-146a by hepatitis B virus X protein contributes to hepatitis development by downregulating complement factor H. MBio 2015. [Crossref] [PubMed]
- Bandiera S, Pernot S, El Saghire H, et al. Hepatitis C Virus-Induced Upregulation of MicroRNA miR-146a-5p in Hepatocytes Promotes Viral Infection and Deregulates Metabolic Pathways Associated with Liver Disease Pathogenesis. J Virol 2016;90:6387-400. [Crossref] [PubMed]
- Fu XY, Tan DM, Liu CM, et al. Early hepatitis B viral DNA clearance predicts treatment response at week 96. World J Gastroenterol 2017;23:2978-86. [Crossref] [PubMed]
- Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 2008;3:375-87. [Crossref] [PubMed]
- Fu X, Tan D, Dou X, et al. A multi-center clinical study comparing Sansure Magb and CAP/CTM HBV tests in the quantitative detection of HBV DNA. J Infect Dev Ctries 2016;10:755-61. [Crossref] [PubMed]
- Wu Z, Han M, Chen T, et al. Acute liver failure: mechanisms of immune-mediated liver injury. Liver Int 2010;30:782-94. [Crossref] [PubMed]
- Wang S, Zhang X, Ju Y, et al. MicroRNA-146a feedback suppresses T cell immune function by targeting Stat1 in patients with chronic hepatitis B. J Immunol 2013;191:293-301. [Crossref] [PubMed]
- Hayes CN, Akamatsu S, Tsuge M, et al. Hepatitis B virus-specific miRNAs and Argonaute2 play a role in the viral life cycle. PLoS One 2012;7:e47490. [Crossref] [PubMed]
- Ma K, He Y, Zhang H, et al. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J Biol Chem 2012;287:5639-49. [Crossref] [PubMed]
- Perry MM, Moschos SA, Williams AE, et al. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol 2008;180:5689-98. [Crossref] [PubMed]
- Flink HJ, van Zonneveld M, Hansen BE, et al. Treatment with Peg-interferon alpha-2b for HBeAg-positive chronic hepatitis B: HBsAg loss is associated with HBV genotype. Am J Gastroenterol 2006;101:297-303. [Crossref] [PubMed]
- Schaefer S. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol 2007;13:14-21. [Crossref] [PubMed]
- Bartholomeusz A, Schaefer S. Hepatitis B virus genotypes: comparison of genotyping methods. Rev Med Virol 2004;14:3-16. [Crossref] [PubMed]


