
The alterations of gut microbiota in mice with chronic pancreatitis
Introduction
Chronic pancreatitis (CP) is characterized by progressive inflammatory and fibrotic changes with the destruction of pancreatic structures, leading to impaired functions of exocrine and endocrine (1). Patients with CP develop the clinical features of abdominal pain, pancreatic exocrine insufficiency (PEI) and diabetes, which have a severely effect on the quality of life (2). The PEI as exogenous enzymes [pancreatic enzyme replacement therapy (PERT)] may reduce bloating, diarrhea, malabsorption, and malnutrition (3-6). However, regardless of the adequacy of PERT, CP patients still have clinical symptoms such as steatorrhea (7). Small intestinal bacterial overgrowth (SIBO) is common in patients with CP, and it can result in chronic intestinal symptoms, including abdominal pain, flatulence, and malabsorption (8,9). Therefore, SIBO should be a potential target for the treatment in CP with PEI.
According to the results of duodenal aspiration cultures, SIBO is typically defined as the overload 105 colony forming units per milliliter (CFU/mL) of bacteria in usually colonic enterobacteria (10). It is suggested that SIBO may aggravate PEI, and SIBO treatment may be helpful for patients with PEI (7). Moreover, studies have shown that oral antibiotic therapy effectively reduced intestinal bacterial overgrowth in dogs with PEI (11) and alleviated intestinal symptoms in SIBO patients (12). In recent years, gut microbiome plays a vital role in human health and disease. Dysbiosis has been found to be related to the activation of inflammatory cytokines in inflammatory-mediated metabolic disease (13), as well as other diseases such as obesity, metabolic syndrome, and diabetes (14-16). However, a few studies have examined links between alterations of intestinal microbiota and CP (17). Therefore, we aim to investigate the variations in diversity, composition and function of intestinal microbiota in CP mice.
Methods
Chemicals and reagents
Cerulein, the decapeptide analogue of cholecystokinin, was purchased from Bachem AG (Bubendorf, Switzerland). All efforts were made to minimize animal suffering. All studies were approved by the Animal Experimental Ethical Inspection Form of Southeast University.
Animals and experimental design
Specific pathogen-free (aged 6–8 weeks) male wild-type C57BL/6 mice were purchased from Animal Laboratory of Nanjing Medical University (Nanjing, China). In the adaptation period (7 days), mice were randomly divided into normal control (CT) and experimental groups (n=8 mice per group) and maintained in a 12 h light/12 h dark cycle at 23±1 °C, with free access to sterilized water and food. For experimental group, mice received single intraperitoneal (IP) injection of alcohol (30%, 3 g/kg/day) 6 days a week for 6 weeks. In addition, the mice were given IP of cerulein every hour for 6 hours (50 µg/kg/body weight) on once a week of 6 consecutive weeks (CP group). Meanwhile, normal CT mice received comparable injections of normal saline instead of cerulein and ethanol for 6 weeks (CT group). The body weights of all mice were recorded at the end of each week and ahead of the final sacrifice. After 6 weeks of ethanol + cerulein or saline administration, the mice were sacrificed and their pancreas and colon feces were collected and stored in –80 °C refrigerator (DWHL-528S, Meiling, Hefei, China).
Evaluation of pancreas morphology
To make paraffin section, fresh pancreas were fixed in 4% paraformaldehyde, paraffin-embedded and sectioned at 3–5 µm. The sections of the specimens were stained with hematoxylin and eosin (HE) and Masson staining according to standard histological examination, then the morphological changes and fibrosis were observed under a light microscope.
DNA extraction and 16S rRNA gene sequencing
Total bacterial genomic DNA samples were extracted using the Metagenomic DNA (MoBio Laboratories, Carlsbad, CA, USA). The quantity and quality of extracted DNAs were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively. The 16S rRNA gene comprising V4 region was amplified by polymerase chain reaction (PCR) using composite specific bacterial primers [the forward primer 515F (5'- GTGCCAGCMGCCGCGGTAA-3') and the reverse primer 806R (5'-GGACTACHVGGGTWTCTAAT-3']. Thermal cycling consisted of the following condition: 98 °C for 30 s (1 cycle), 98 °C for 15 s/58 °C for 15 s/72 °C for 15 s (30 cycles), and a final extension at 72 °C for 1 min. High-throughput pyrosequencing of the PCR products was performed on an Illlumina HiSeq4000 platform at GUHE Info technology Co., Ltd (Hangzhou, China).
Gut microbiota sequence analysis
Sequence data analyses were mainly performed using Quantitative Insights Into Microbial Ecology (QIIME) and R packages (v3.2.0). The raw paired-end reads from the original DNA fragments were merged using FLASH (18), and assigned to each sample according to the unique barcodes. High-quality reads for bioinformatics analysis were performed and all of the effective reads from each sample were clustered into operational taxonomic units (OTUs) based on a 97% sequence similarity according to VSEARCH (19). OTU-level alpha diversity indices, such as Chao1 richness estimator, Shannon diversity index, and Simpson index, were calculated using the OTU table in QIIME (20). OTU-level ranked abundance curves were generated to compare the richness and evenness of OTUs among samples. Beta diversity analysis was performed to investigate the structural variation of microbial communities across samples using UniFrac distance metrics (21,22), and visualized via principal coordinate analysis (PCoA) and principal component analysis (PCA) (23). Linear discriminant analysis effect size (LEfSe) was performed to detect differentially abundant taxa across groups using the default parameters (24). BugBase (http://github.com/danknights/bugbase) was used to calculate differences between both groups in terms of microbial phenotypes, based on high-quality sequences (25). The output file was further analysed using statistical analysis of metagenomic profiles (STAMP) software package v2.1.3 (26).
Statistical analysis
Data were expressed as mean ± standard deviation (SD), and the differences between groups with normal distribution were evaluated by the Student’s t-test using SPSS 19.0. The criterion of significance was conducted at P<0.05.
Results
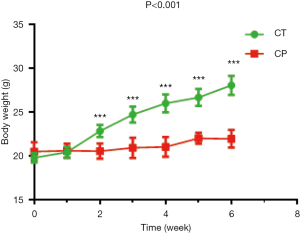
Body weight
The weight of mice in the experimental group was significantly lower than CT group from the second week, as shown in Figure 1. After 6 weeks, the average body weight of the mice was 21.96±1.00 g in CP group significantly lower than CT group (28.05±1.07 g) (P<0.001).
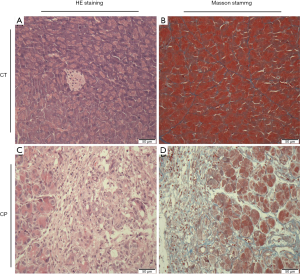
Pancreatic histopathological assessment
As shown in Figure 2A,B, the HE staining of the pancreas tissues revealed profound acinar loss, infiltration of inflammatory cells, enlarged interstitial spaces, glandular atrophy, degeneration and parenchyma fibrosis in the CP group. Masson staining (Figure 2C,D) showed extensive trichrome-positive staining in the CP group.
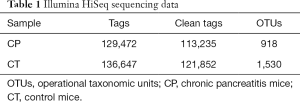
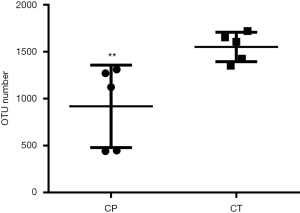
Generation of OTUs
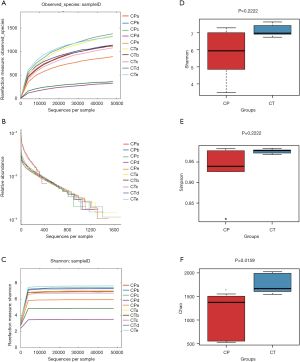
For the two groups, variable regions (V4) of the bacterial 16S rRNA gene were amplified by PCR. The raw reads of high-throughput pyrosequencing in CP and CT groups were 129,472 and 136,647, respectively. After removing the low-quality sequences, the two groups of clean tags were 113,235 and 121,852, respectively, and were subjected to the following analysis. The CP and CT groups were identified with 918 and 1,530 OTUs, respectively, based on the conventional criterion of 97% similarity (equal to species level) (Table 1). The average length of the sequences was 2 bp × 150 bp. Moreover, there was significant differences in the number of OTUs between CP group and CT group in Figure 3.
Full table
Gut microbial dysbiosis in CP mice
Richness and diversity of bacteria phylotypes
The curves of OTU rank (Figure 4A), rarefaction (Figure 4B), Shannon-Winner curves (Figure 4C), indexes of Shannon (Figure 4D), Simpson (Figure 4E), and Chao1 (Figure 4F) were calculated. Cerulein combined with alcohol significantly reduced the richness of the intestinal microbiota in CP mice.
Bacterial taxonomic composition
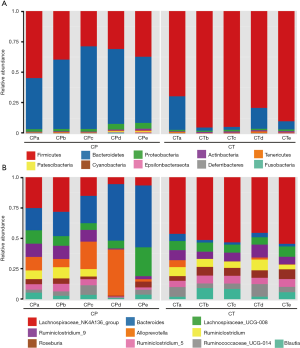
The sequences were analyzed at multiple (phylum to genus) taxonomic levels. At the phylum levels, gut microbiota was mainly composed of Firmicutes, Bacteroidetes, Proteobacteria among the two groups. The relative abundance of Bacteroidetes (55.8% vs. 11.2%) significantly increased and the relative abundance of Firmicutes (38.0% vs. 85.5%) decreased in CP group compared with the CT group (Figure 5A), thereby reducing the ratio of Firmicutes/Bacteroidetes. At the genus levels, the CP group had higher abundance of bacteria like Bacteroides and Alloprevotella, and lower significantly such as Lachnospiraceae_NK4A136, Ruminiclostridium, Roseburia and Blautia compared to the CT group (P<0.05 all) (Figure 5B).
Taxonomic characterization of the gut microbial profile
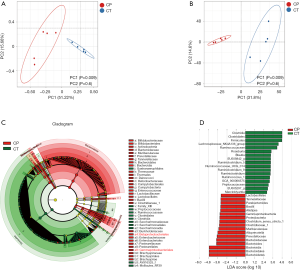
Weighted UniFrac cluster tree based on UPGMA showed the beta diversity changes of intestinal flora in the two groups (Figure 6A,B). LEfSe was used to identify differential microbial abundances between CP and healthy mice. This analysis revealed significant differences in bacterial clades from phylum to species level between groups, e.g., Lachnospiraceae and Ruminococcaceae and Roseburia were more abundant in the CT group, whereas the families Bacteroidaceae, Muribaculaceae, Prevotellaceae, Enterobacteriaceae and Bifidobacteriaceae were more abundant in the CP mice (Figure 6C,D).
Microbial potential functions
Using BugBase, nine potential phenotypes, including aerobic, anaerobic, containing mobile elements, facultatively anaerobic, biofilm forming, gram-negative, gram-positive, potentially pathogenic, and stress tolerant were predicted significant in the two groups (P<0.05), as shown Table 2. Among all the phenotypes, CP mice tend to have more aerobic, facultatively anaerobic and biofilm forming, and less anaerobic and containing mobile elements bacteria. Additionally, we observed that a significantly lower representation of gram-positive bacteria, whereas proportion of gram-negative and potentially pathogenic phenotypes were observed to be significantly enriched in the CP mice.

Full table
Discussion
There is a lack of knowledge in regard to the changes in diversity, composition and function of the gut microbiome in mice with CP. In this current study, we found that the CP mice indicated reduced weight gain and fibrogenesis processes of pancreas. Mice with CP had a distinct gut microbiota profile compared to the healthy mice. Moreover, we first explored the microbial community level phenotypes, revealing that the proportion of gram-negative and potentially pathogenic phenotypes was increased in mice with CP.
Alcohol intake is the main etiology factor related to CP (27). It is generally suggested that CP may be caused by repeated acute pancreatitis (28). The combination of alcohol and cerulein leads to typical pancreatic fibrosis, activation of stellate cells, and inflammation of the pancreas (29). In our study, the repeated injection with ethanol and caerulein for 6 weeks caused CP. All three main pathological criteria of human CP, including parenchyma fibrosis, loss of acinar cells, and infiltration of inflammatory cells, were observed in present CP model mice.
Alpha diversity has been shown to be potential associated to the pathogenesis in various diseases. For example, a lower bacterial diversity is related to inflammatory bowel disease (30). Our results show the non-significant difference in bacterial alpha diversity (Shannon and Simpson index) in the two groups, whereas CP mice had the lower species richness (observed OTUs and Chao1) than the healthy mice. Therefore, the decrease of bacterial diversity in CP mice may promote the progress of the disease.
Moreover, our findings showed that the intestinal microbial community of mice with CP was significantly different from normal mice. Previous studies have shown that the gut microbial community of Crohn’s disease patients was characterized by decrease in phylum Firmicutes and concomitant increase in phylum Proteobacteria (31,32). Among the top 3 predominant, the abundance of Firmicutes was decreased, Bacteroidetes and Proteobacteria were more abundant in CP group, which was consistent with previous studies (33). However, inconsistent results have been reported. Hu et al. found (34,35) that the proportion of Bacteroidetes was decreased with CP mice, an increased proportion of Firmicutes was observed in study. Multiple bacteria of the phylum Firmicutes was significantly decreased in the gut microbial community of Crohn’s disease patients (36). Indeed, the ratio of Firmicutes/Bacteroidetes has been used as a significant indicator to estimate the status of human gut microbes (37,38). Our study revealed a decreased ratio of Firmicutes/Bacteroidetes in CP mice, suggesting that the progression of CP might result in the disproportions of Firmicutes and Bacteroidetes in the gut.
At the genus levels, CP mice reduced the abundance of butyrate-producing bacterial species, and several anti-inflammatory bacteria including Lachnospiraceae_NK4A136, Ruminococcus, Roseburia and Blautia, and these taxa have been considered as beneficial microbiota for gut health (39-41). Recently, Jandhyala et al. (35,42) have evaluated that the Ruminococcus abundance of gut microbiome was reduced in patients with CP. Moreover, our study found that conditional pathogenic bacteria was increased in CP mice, including Bacteroides and Enterobacteriales. Previously, increased conditional pathogenic bacteria has been reported in fecal extracts of Crohn’s disease patients (36,43). In contrast, other beneficial bacteria such as Lactobacillus, Alloprevotella, and Bifidobacterium were significantly increased in the CP mice, nevertheless, the relevance of these changes was unclear. Some studies have been reported higher prevalence of Lactobacillus and Bifidobacterium in gastrointestinal disorders (44). Consequently, the differences in beta diversity were found to be due to significant decreases in prominent members of the gut microbiota with a significant increase in mice with CP.
At the microbial community level, gene functions related to aerobic, facultative anaerobiosis, and form biofilms were increased in the CP group. Anaerobic and gram-positive species were significantly depleted in CP mice, likely owing to the decreased abundance of Lachnospiraceae and Ruminococcaceae. CP is a disease characterized by maldigestion with malabsorption and malnutrition. Million et al. (45) found a depletion of anaerobes in malnutrition. In additional, significant enrichment of gram-negative and potential pathogenic bacteria were predicted for the group, which were majorly attributed to increase the abundance of Bacteroidetes. Therefore, the intestinal microflora of CP mice was changed compared with that of healthy mice.
There are some limitations in our study. First, we evaluated only a small number of animals in the CP group, limiting our ability to fully characterize the microbiome of CP mice. Second, we did not consider to evaluate samples of the small intestine. Third, it may be more valuable to observe the time course of changes in intestinal microflora by the development of CP. Finally, there is no evidence for the causal relationship between CP and intestinal flora imbalance.
Conclusions
This study indicated that mice with CP had a distinct microbiota profile.
Acknowledgments
Funding: This work was funded by the National Natural Science Foundation of China (No. 81570739 and No. 81800571).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All efforts were made to minimize animal suffering. All studies were approved by the Animal Experimental Ethical Inspection Form of Southeast University (No. 20190218001).
References
- Majumder S, Chari ST. Chronic pancreatitis. Lancet 2016;387:1957-66. [Crossref] [PubMed]
- Lévy P, Domínguez-Muñoz E, Imrie C, et al. Epidemiology of chronic pancreatitis: burden of the disease and consequences. United European Gastroenterol J 2014;2:345-54. [Crossref] [PubMed]
- de la Iglesia-García D, Huang W, Szatmary P, et al. Efficacy of pancreatic enzyme replacement therapy in chronic pancreatitis: systematic review and meta-analysis. Gut 2017;66:1354-5. [Crossref] [PubMed]
- Thorat V, Reddy N, Bhatia S, et al. Randomised clinical trial: the efficacy and safety of pancreatin enteric-coated minimicrospheres (Creon 40000 MMS) in patients with pancreatic exocrine insufficiency due to chronic pancreatitis—a double-blind, placebo-controlled study. Aliment Pharmacol Ther 2012;36:426-36. [Crossref] [PubMed]
- Whitcomb DC, Lehman GA, Vasileva G, et al. Pancrelipase delayed-release capsules (CREON) for exocrine pancreatic insufficiency due to chronic pancreatitis or pancreatic surgery: a double-blind randomized trial. Am J Gastroenterol 2010;105:2276-86. [Crossref] [PubMed]
- Martínez J, Abad-González A, Aparicio JR, et al. The Spanish Pancreatic Club recommendations for the diagnosis and treatment of chronic pancreatitis: part 1 (diagnosis). Pancreatology 2013;13:8-17. [Crossref] [PubMed]
- Ní Chonchubhair HM, Bashir Y, Dobson M, et al. The prevalence of small intestinal bacterial overgrowth in non-surgical patients with chronic pancreatitis and pancreatic exocrine insufficiency (PEI). Pancreatology 2018;18:379-85. [Crossref] [PubMed]
- Capurso G, Signoretti M, Archibugi L, et al. Systematic review and meta-analysis: small intestinal bacterial overgrowth in chronic pancreatitis. United European Gastroenterol J 2016;4:697-705. [Crossref] [PubMed]
- Kim DB, Paik CN, Sung HJ, et al. Breath hydrogen and methane are associated with intestinal symptoms in patients with chronic pancreatitis. Pancreatology 2015;15:514-8. [Crossref] [PubMed]
- Choung RS, Ruff KC, Malhotra A, et al. Clinical predictors of small intestinal bacterial overgrowth by duodenal aspirate culture. Aliment Pharmacol Ther 2011;33:1059-67. [Crossref] [PubMed]
- Westermarck E, Myllys V, Aho M. Effect of treatment on the jejunal and colonic bacterial flora of dogs with exocrine pancreatic insufficiency. Pancreas 1993;8:559-62. [Crossref] [PubMed]
- Shah SC, Day LW, Somsouk M, et al. Meta-analysis: antibiotic therapy for small intestinal bacterial overgrowth. Aliment Pharmacol Ther 2013;38:925-34. [Crossref] [PubMed]
- Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology 2017;152:327-39.e4. [Crossref] [PubMed]
- West-Eberhard MJ. Nutrition, the visceral immune system, and the evolutionary origins of pathogenic obesity. Proc Natl Acad Sci U S A 2019;116:723-31. [Crossref] [PubMed]
- Org E, Blum Y, Kasela S, et al. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol 2017;18:70. [Crossref] [PubMed]
- Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012;490:55-60. [Crossref] [PubMed]
- Memba R, Duggan SN, Ni Chonchubhair HM, et al. The potential role of gut microbiota in pancreatic disease: a systematic review. Pancreatology 2017;17:867-74. [Crossref] [PubMed]
- Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011;27:2957-63. [Crossref] [PubMed]
- Rognes T, Flouri T, Nichols B, et al. VSEARCH: a versatile open source tool for metagenomics. PeerJ 2016;4:e2584. [Crossref] [PubMed]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335-6. [Crossref] [PubMed]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 2005;71:8228-35. [Crossref] [PubMed]
- Lozupone CA, Hamady M, Kelley ST, et al. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 2007;73:1576-85. [Crossref] [PubMed]
- Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 2007;62:142-60. [Crossref] [PubMed]
- Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [Crossref] [PubMed]
- Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31:814-21. [Crossref] [PubMed]
- Parks DH, Tyson GW, Hugenholtz P, et al. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 2014;30:3123-4. [Crossref] [PubMed]
- Pandol SJ, Raraty M. Pathobiology of alcoholic pancreatitis. Pancreatology 2007;7:105-14. [Crossref] [PubMed]
- Magnusdottir BA, Baldursdottir MB, Kalaitzakis E, et al. Risk factors for chronic and recurrent pancreatitis after first attack of acute pancreatitis. Scand J Gastroenterol 2019;54:87-94. [Crossref] [PubMed]
- Charrier AL, Brigstock DR. Connective tissue growth factor production by activated pancreatic stellate cells in mouse alcoholic chronic pancreatitis. Lab Invest 2010;90:1179-88. [Crossref] [PubMed]
- Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology 2008;134:577-94. [Crossref] [PubMed]
- Mizoguchi A, Mizoguchi E. Inflammatory bowel disease, past, present and future: lessons from animal models. J Gastroenterol 2008;43:1-17. [Crossref] [PubMed]
- Nagalingam NA, Lynch SV. Role of the microbiota in inflammatory bowel diseases. Inflamm Bowel Dis 2012;18:968-84. [Crossref] [PubMed]
- Li K, Zhuo C, Teng C, et al. Effects of Ganoderma lucidum polysaccharides on chronic pancreatitis and intestinal microbiota in mice. Int J Biol Macromol 2016;93:904-12. [Crossref] [PubMed]
- Hu Y, Teng C, Yu S, et al. Inonotus obliquus polysaccharide regulates gut microbiota of chronic pancreatitis in mice. AMB Express 2017;7:39. [Crossref] [PubMed]
- Jandhyala SM, Madhulika A, Deepika G, et al. Altered intestinal microbiota in patients with chronic pancreatitis: implications in diabetes and metabolic abnormalities. Sci Rep 2017;7:43640. [Crossref] [PubMed]
- Takahashi K, Nishida A, Fujimoto T, et al. Reduced abundance of butyrate-producing bacteria species in the fecal microbial community in Crohn’s disease. Digestion 2016;93:59-65. [Crossref] [PubMed]
- Mariat D, Firmesse O, Levenez F, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 2009;9:123. [Crossref] [PubMed]
- Indiani CMDSP, Rizzardi KF, Castelo PM, et al. Childhood obesity and Firmicutes/Bacteroidetes ratio in the gut microbiota: a systematic review. Child Obes 2018;14:501-9. [Crossref] [PubMed]
- Walter J. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol 2008;74:4985-96. [Crossref] [PubMed]
- Meehan CJ, Beiko RG. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol 2014;6:703-13. [Crossref] [PubMed]
- Huang G, Zhang S, Zhou C, et al. Influence of Eimeria falciformis infection on gut microbiota and metabolic pathways in mice. Infect Immun 2018;86. [Crossref] [PubMed]
- Isaiah A, Parambeth JC, Steiner JM, et al. The fecal microbiome of dogs with exocrine pancreatic insufficiency. Anaerobe 2017;45:50-8. [Crossref] [PubMed]
- Andoh A, Tsujikawa T, Sasaki M, et al. Faecal microbiota profile of Crohn’s disease determined by terminal restriction fragment length polymorphism analysis. Aliment Pharmacol Ther 2009;29:75-82. [Crossref] [PubMed]
- Wang W, Chen L, Zhou R, et al. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol 2014;52:398-406. [Crossref] [PubMed]
- Million M, Tidjani Alou M, Khelaifia S, et al. Increased gut redox and depletion of anaerobic and methanogenic prokaryotes in severe acute malnutrition. Sci Rep 2016;6:26051. [Crossref] [PubMed]


