
Hes1 is associated with long non-coding RNAs in colorectal cancer
Introduction
Among the most commonly diagnosed cancers, colorectal cancer (CRC) is a major cause of morbidity and mortality throughout the world. It is practical to clarify the molecular mechanisms underlying CRC carcinogenesis and progression.
Long noncoding RNAs (lncRNAs) are transcripts with a length greater than 200 nt and devoid of coding potential of functional proteins (1). According to the localization of lncRNAs relative to protein-coding target mRNAs, lncRNAs have been classified as intronic lncRNAs, antisense lncRNAs, long intergenic ncRNAs (lincRNAs), enhancer RNAs (eRNAs), or transcribed pseudogene lncRNAs (2). Studies have demonstrated that lncRNAs play integral roles in the control of cellular growth, division and differentiation by regulating downstream target genes at an epigenetic, transcription, or posttranscriptional level (3,4). The perturbation of lncRNA expression can contribute to the development and progression of cancer including CRC (5-7). LncRNAs have been shown to play a crucial role in the development as well as in the prognosis of various human cancers. LncRNAs participate in various biological processes (such as genomic imprinting, chromatin modeling and post-transcriptional regulation), it is presumed that dysregulation of lncRNA expression could hamper cellular homeostasis and lead to cancer initiation (8). Over the past decade, multiple studies have identified alterations in lncRNA expression patterns in the context of cancers, potentially resulting from genetic and epigenetic changes including chromosomal translocations, copy number alterations, small insertions and deletions (INDELs) and single nucleotide polymorphisms (SNP) (9,10) or dysregulation by specific oncogenic and tumor-suppressor related signals and regulatory factors (11-13).
Currently, lncRNAs have been demonstrated to interact with RNA-binding proteins to regulate protein function, participate in chromatin remodeling and histone modification, and function as competing endogenous RNA (ceRNA) (8).
Hes1 is a transcription factor that influences cell proliferation and differentiation in embryogenesis. Our previous study showed that Hes1 was involved in the self-renewal and tumorigenicity of stem-like cancer cells in CRC (14) .
In this study, we firstly detected considerable up-regulated and down-regulated lncRNAs using ArrayStar Human LncRNA/mRNA Expression Microarray Version 3.0. Then we screened a significant number of lncRNAs interacting with Hes1 by RIP-seq assay. Interestingly, we found several lncRNAs were identical to those in the tissue microarray. Next, we verified the binding of these lncRNAs to Hes1 and the expression of the lncRNAs in CRC samples. These above-mentioned findings and information encouraged us to fully investigate whether these lncRNAs play important roles in the development and progression of CRC.
Methods
Clinical samples and ethics statement
The biopsies of 32 CRC patients and 32 non-tumoral tissues were collected from The First Affiliated Hospital of Jinan University and Nanfang Hospital of Guangzhou City in China. None of the patients received preoperative radiotherapy or chemotherapy. Approval from the Jinan University Institute Research Ethics Committee was obtained. The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
RNA isolation, reverse transcription, and qRT-PCR
For the mRNA analyses, the total RNA was extracted using the Trizol Reagent (Invitrogen) according to the protocol provided by the manufacturer. The total RNA was reversely transcribed using the PrimeScript RT reagent Kit (TaKaRa). The qRT-PCR for the analysis of mRNA expression was performed on a Stratagene Mx3005P qRT-PCR system using the SYBR Green qRT-PCR master mix (TaKaRa) and GAPDH for normalisation. The primers used for the amplification of the indicated genes are listed in Table S1. All of the samples were normalised to the internal controls, and the fold changes were calculated through relative quantification (2−ΔΔCt).

Full table
Microarray analysis
The total RNA was extracted from normal colorectal tissues and tumor tissues using TRIzol® Reagent (Invitrogen, CA, USA). Gene expression was determined using The ArrayStar Human LncRNA/mRNA Expression Microarray Version 3.0 (ArrayStar, MD, USA) following the manufacturer’s instructions. Acquired array images were analyzed using the Agilent Feature Extraction Software. Differentially expressed lncRNAs and mRNAs were identified by setting the cutoff for P values corrected for multiple comparisons by the Benjamini Hochberg false discovery rate method at Q <0.05 and for fold changes at >2 (up- or down-regulated).
RNA-binding protein immunoprecipitation (RIP)
We carried out RIP using Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, MD, USA) following the manufacturer’s instructions. Cells were lysed in RIP Lysis Buffer, and immunoprecipitated with Hes1 antibody (CST, MA, USA) to RNA-binding protein of interest with protein A/G magnetic beads. RNAs were extracted and analyzed by RT-PCR or Illumina HiSeq 2500 Sequencing.
RNA pull-down assay
LncRNAs were in vitro transcribed respectively from vector pGM-T Easy (Promega #A3600, WI, USA), and biotin-labeled with the Biotin RNA Labeling Mix (Roche, CA, USA) and T7 RNA polymerase (Roche), treated with RNase-free DNase I (Roche, CA, USA), and purified with a RNeasy Mini Kit (Qiagen, MD, USA). One milligram of whole-cell lysates from SW480 cells were incubated with 3 µg of purified biotinylated transcripts for 1 h at 25 °C; complexes were isolated with streptavidin agarose beads (Invitrogen, CA, USA). Beads were washed briefly 5 times in Handee spin columns (Pierce, WI, USA) and boiled in SDS buffer, and the retrieved protein was detected by standard Western blot technique using Hes1 antibody (CST, MA, USA).
Results
LncRNAs expression in CRC
In order to study the roles of lncRNAs in CRC, we first detected lncRNA expression in three CRC tissues and three non-tumoral tissues (two patients were poorly differentiated CRC and one patient was well and moderately differentiated CRC confirmed by pathology) using ArrayStar Human LncRNA/mRNA Expression Microarray Version 3.0. Our results demonstrated significantly up-regulated and down-regulated lncRNAs (Figure 1; GEO series: GSE134552) in CRC tissues compared with non-tumoral tissues.
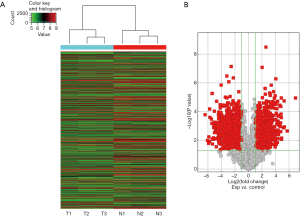
Hes1 is associated with lncRNAs in CRC
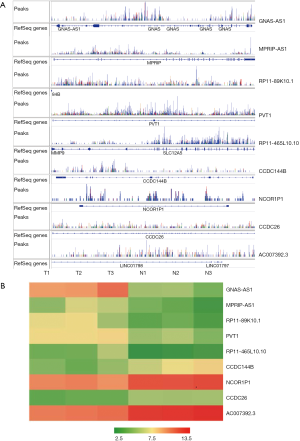
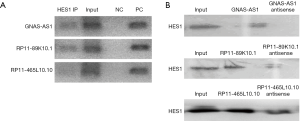
Since primarily we have reported that Hes1 is involved in the self-renewal and tumorigenicity of stem-like cancer cells in CRC, to detect if there are any lncRNAs binding to Hes1, we performed RIP-seq using Hes1 antibody. Interestingly, we found that Hes could bind several lncRNAs (Figure 2A; GEO series: GSE134552), and lncRNAs such as GNAS-AS1, MPRIP-AS1, RP11-89K10.1, PVT1, and PR-11-465L10.10 were up-regulated, while CCDC144B, NCOR1P1, CCDC26, and AC007392.3 were down-regulated according to the LncRNA/mRNA Expression Microarray (Figure 2B). Because the expression of Hes1 in poorly differentiated CRC samples is higher than that observed in well-differentiated tumor as we reported before (14), next, we detected the direct bindings between those picked up-regulated lncRNAs (GNAS-AS1, RP11-89K10.1, RP11-465L10.10) and Hes1 separately by doing RIP-PCR using Hes1 antibody (Figure 3A) and RNA Pulldown using lncRNAs and their antisense sequences (Figure 3B).
Up-regulated lncRNAs binding to Hes1 in enlarged CRC tissue samples
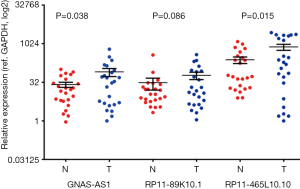
To confirm the expression of these Hes1 related lncRNAs (GNAS-AS1, RP11-89K10.1, RP11-465L10.10), we detected their expression in 32 more CRC samples by real-time PCR. We found that GNAS-AS1 and RP11-465L10.10 were significantly up-regulated in 32 CRC samples compared with the adjacent non-tumoral tissues (P<0.05), RP11-89K10.1 expression also showed a robust up-regulation in CRC tissues although the results were not statistically significant (Figure 4), indicating that these lncRNAs play important roles in the development and progression of CRC, and modulates biological functions of CRC via interacting with Hes1.
Discussion
The aberrant lncRNA expression has been observed in many cancers, and cancer-associated lncRNAs display aberrant expression patterns in tissue-type specific manner, indicating their potentials as biomarkers for cancer diagnosis and prognosis (15-18). The mechanisms by which lncRNAs exert their functions require elucidation to optimize their potential utility as alternative biomarkers and therapeutic targets during tumor occurrence and progression.
The Notch1 signaling pathway plays a vital role in tumorigenesis of lung cancer cells. Baumgart et al. showed that activation of Notch1 resulted in regulation of epidermal growth factor receptor (EGFR) expression by ADAM17 during tumorigenicity of non-small cell lung cancer (NSCLC) cells (19). Ji et al. reported that induction of miR-34a resulted in inhibition of proliferation and invasion in NSCLC via the Notch-1 pathway (20). Our previous study showed that Hes1 was significantly up-regulated in poorly differentiated CRC samples. Moreover, Hes1 was involved in the self-renewal and tumorigenicity of stem-like cancer cells in CRC, and promotes cell proliferation and migration by activating Bmi-1 and PTEN/Akt/GSK3β pathway in human CRC (14,21).
In this study, we firstly screened considerable up-regulated and down-regulated lncRNAs using Phalanx human OneArray microarray. These lncRNAs may represent potential prognostic markers for predicting the aggressiveness of CRC. Then we screened a significant number of lncRNAs interacting with Hes1 by RIP-sequence assay. Interestingly, we found several lncRNAs were identical to those in the tissue microarray. GNAS is an imprinted locus that produces four monoallelic (NESP55, GNAS-AS1, XLsα, and A/B) transcripts due to differential methylation of their promoters (DMR) in human pluripotent stem cells, and it has been reported that GNAS-AS1/miR-4319/NECAB3 axis promotes migration and invasion of NSCLC cells by altering macrophage polarization (22,23). RP11-89K10.1 was reported to be involved in chimeras in acute myeloid leukemia (24). And from bioinformatics prediction, it’s possible that RP11-89K10.1 plays functions via hsa-miR-22-5p (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted&miRNAs%5B0%5D=hsa-miR-22-5p&page=18). Researchers found that there was a strong RNA-protein interaction between RP11-465L10.10 and myeloid zinc finger gene 1 (MZF1), a transcription factor of MMP9, and may play important roles in thoracic aortic aneurysm. Next, we verified that Hes1 could directly interact with lncRNAs (GNAS-AS1, RP11-89K10.1, RP11-465L10.10) by RIP and RNA pull down assays. Finally, we detected the expression of them in CRC samples by real-time PCR. Based on these, we speculated that Hes1 may interact with one or more lncRNAs which play a quantitative role in the development and progression of CRC.
Furthermore, cancer-specific lncRNA expression patterns appear more tissue- and stage-specific than those of protein-coding genes, supporting the potential development of lncRNAs as powerful alternative biomarkers and therapeutic targets (25,26).
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of Guangdong Province, China (grant No. 2018A030310604 and grant No. 2017A030310081) and Pearl River Nova Program, Science and Technology Program of Guangzhou, China (grant No. 201710010092).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Approval from the Jinan University Institute Research Ethics Committee was obtained (No. 20170225).
References
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015;21:1253-61. [Crossref] [PubMed]
- Heward JA, Roux BT, Lindsay MA. Divergent signalling pathways regulate lipopolysaccharide-induced eRNA expression in human monocytic THP1 cells. FEBS Lett 2015;589:396-406. [Crossref] [PubMed]
- Shin TJ, Lee KH, Cho HM, et al. Concise approach for screening long non-coding RNAs functionally linked to human breast cancer associated genes. Exp Mol Pathol 2019;108:89-96. [Crossref] [PubMed]
- Gao Z, Chen M, Tian X, et al. A novel human lncRNA SANT1 cis-regulates the expression of SLC47A2 by altering SFPQ/E2F1/HDAC1 binding to the promoter region in renal cell carcinoma. Rna Biol 2019;16:940-9. [Crossref] [PubMed]
- Diao P, Ge H, Song Y, et al. Overexpression of ZEB2-AS1 promotes epithelial-to-mesenchymal transition and metastasis by stabilizing ZEB2 mRNA in head neck squamous cell carcinoma. J Cell Mol Med 2019;23:4269-80. [Crossref] [PubMed]
- Cheng D, Fan J, Ma Y, et al. LncRNA SNHG7 promotes pancreatic cancer proliferation through ID4 by sponging miR-342-3p. Cell Biosci 2019;9:28. [Crossref] [PubMed]
- Li M, Bian Z, Jin G, et al. LncRNA-SNHG15 enhances cell proliferation in colorectal cancer by inhibiting miR-338-3p. Cancer Med 2019;8:2404-13. [Crossref] [PubMed]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904-14. [Crossref] [PubMed]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol 2011;21:354-61. [Crossref] [PubMed]
- Du Z, Fei T, Verhaak RG, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol 2013;20:908-13. [Crossref] [PubMed]
- Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer 2011;129:773-9. [Crossref] [PubMed]
- Nakano S, Murakami K, Meguro M, et al. Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Cancer Sci 2006;97:1147-54. [Crossref] [PubMed]
- Weksberg R, Nishikawa J, Caluseriu O, et al. Tumor development in the Beckwith-Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Hum Mol Genet 2001;10:2989-3000. [Crossref] [PubMed]
- Gao F, Zhang Y, Wang S, et al. Hes1 is involved in the self-renewal and tumourigenicity of stem-like cancer cells in colon cancer. Sci Rep 2014;4:3963. [Crossref] [PubMed]
- Bolha L, Ravnik-Glavač M, Glavač D. Long Noncoding RNAs as Biomarkers in Cancer. Dis Markers 2017;2017:7243968.
- Mouraviev V, Lee B, Patel V, et al. Clinical prospects of long noncoding RNAs as novel biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostatic Dis 2016;19:14-20. [Crossref] [PubMed]
- Yang Z, Guo X, Li G, et al. Long noncoding RNAs as potential biomarkers in gastric cancer: Opportunities and challenges. Cancer Lett 2016;371:62-70. [Crossref] [PubMed]
- Zhong Y, Wu X, Li Q, et al. Long noncoding RNAs as potential biomarkers and therapeutic targets in gallbladder cancer: a systematic review and meta-analysis. Cancer Cell Int 2019;19:169. [Crossref] [PubMed]
- Baumgart A, Seidl S, Vlachou P, et al. ADAM17 Regulates Epidermal Growth Factor Receptor Expression through the Activation of Notch1 in Non-Small Cell Lung Cancer. Cancer Res 2010;70:5368-78. [Crossref] [PubMed]
- Ji X, Wang Z, Geamanu A, et al. Delta-tocotrienol suppresses Notch-1 pathway by upregulating miR-34a in nonsmall cell lung cancer cells. Int J Cancer 2012;131:2668-77. [Crossref] [PubMed]
- Gao F, Huang W, Zhang YQ, et al. Hes1 promotes cell proliferation and migration by activating Bmi-1 and PTEN/Akt/GSK3 beta pathway in human colon cancer. Oncotarget 2015;6:38667-80. [Crossref] [PubMed]
- Grybek V, Aubry L, Maupetit-Mehouas S, et al. Methylation and Transcripts Expression at the Imprinted GNAS Locus in Human Embryonic and Induced Pluripotent Stem Cells and Their Derivatives. Stem Cell Reports 2014;3:432-43. [Crossref] [PubMed]
- Li Z, Feng C, Guo J, et al. GNAS-AS1/miR-4319/NECAB3 axis promotes migration and invasion of non-small cell lung cancer cells by altering macrophage polarization. Funct Integr Genomics 2019. [Epub ahead of print]. [Crossref] [PubMed]
- L Abbate A, Tolomeo D, Cifola I, et al. MYC-containing amplicons in acute myeloid leukemia: genomic structures, evolution, and transcriptional consequences. Leukemia 2018;32:2152-66. [Crossref] [PubMed]
- Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res 2014;24:616-28. [Crossref] [PubMed]
- Necsulea A, Soumillon M, Warnefors M, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014;505:635-40. [Crossref] [PubMed]


