
Lower detectability of non-invasive prenatal testing compared to prenatal diagnosis in high-risk pregnant women
Introduction
Prenatal screening and diagnosis (1,2) involves an initial serological prenatal screening, after which pregnant women at high risk undergo invasive prenatal diagnostic procedures. The emergence of noninvasive prenatal testing (NIPT) has transformed prenatal screening and diagnosis. However, several problems remain to be solved before widespread clinical application of NIPT.
NIPT detects genetic changes using cell-free DNA (cfDNA) by massively parallel sequencing (MPS). It is currently widely used in prenatal screening for trisomy 21 (T21), trisomy 18 (T18), and trisomy 13 (T13), for which it has satisfactory accuracy (detection rate 99.7%, false positive rate 0.04% for T21; 97.9% and 0.04% for T18; and 99.0% and 0.04% for T13, respectively) (3). NIPT enables detection of not only three common foetal aneuploidies (2,4,5) but also other genetic diseases, such as foetal sex chromosome aneuploidy (1,6) and sub-microscopic chromosomal abnormalities (7).
The consensus opinion on NIPT is that serological prenatal screening should remain the first-line screening method, followed by invasive prenatal diagnostic procedures. NIPT may thus have a role as a second-line screening method. In 2015, the American College of Obstetricians and Gynecologists (8) recommended that NIPT be considered for prenatal screening of foetal aneuploidy but traditional prenatal screening methods should remain the first-line option. The International Society for Prenatal Diagnosis (9) stated that NIPT could be performed in women at intermediate risk after serological prenatal screening to reduce the rate of missed diagnoses. In 2016, the American College of Medical Genetics and Genomics (10) updated the consensus guidelines and recommended that noninvasive prenatal screening (NIPS) could replace traditional serological prenatal screening. In China, perinatal physicians tend to use NIPT as the second-line prenatal screening method. In clinical practice, pregnant women at high risk frequently refuse invasive prenatal diagnostic procedures because of fear of abortion and/or infection. Therefore, the number of pregnant women at high risk willing to undergo NIPT is increasing. However, NIPT was developed to detect T21, T18, T13, and its ability to detect any of the various other foetal chromosomal abnormalities (11) is limited. Therefore, the effect of switching from amniocentesis to NIPT for pregnant women at high risk needs to be evaluated.
Standard karyotype analyses, which can detect major chromosomal abnormities (e.g., aneuploidy, unbalanced rearrangements, Robertsonian translocation, and mosaicism), are typically used for prenatal diagnosis. Recently, chromosomal microarray analysis (CMA), a high resolution genomic technology, has been applied for prenatal diagnosis of genetic disorders. Unlike standard karyotype analyses, CMA can also detect sub-microscopic imbalances or copy number variants (CNVs) (12,13). According to our clinical experience, an increasing number of women are willing to undergo CMA for prenatal diagnosis, particularly in combination with abnormal ultrasonography.
In the present study, we evaluated the rate of diagnosis of chromosomal abnormalities by NIPT compared to invasive prenatal diagnosis in pregnant women at high risk for T21 and T18. We retrospectively explored the results of cytogenetic analyses by amniocentesis in two prenatal diagnosis centres. The results will be used to improve the feasibility of NIPT for prenatal genetic screening.
Methods
Patients and design
From January 2009 to March 2018, 3,099 pregnant women determined to be at high risk by serological prenatal screening at Changzhou Maternity and Child Health Care Hospital (affiliated with Nanjing Medical University) and Lianyungang Maternal and Child Health Hospital (affiliated with Yangzhou Medical University) were enrolled in this study. After genetic counselling, the women underwent prenatal diagnosis by amniocentesis. All of the women had single pregnancies, were 22–38 years of age (mean, 26.7 years), and were at gestational weeks 15+2–20+4 (mean, 16+3 weeks). For pregnant women at high risk of T21 and T18, we evaluated the rate of diagnosis of chromosomal abnormalities by NIPT (Figure 1).
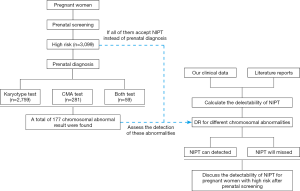
The study design and protocol were reviewed and approved by the Ethics Committee of the Changzhou Maternity and Child Health Care Hospital. Written informed content was obtained from the women prior to screening and diagnosis.
Laboratory methodology
Prenatal serological screening
Serological screening was performed as described previously (2). The concentrations of AFP, free βHCG, and free E3 were determined using a time-resolved immunofluorescence assay. The risks for neural tube defects (NTDs), T21, and T18, taking into account maternal age, gestational age, body weight, and diabetes, were calculated using Lifecycle software (4.0). A high risk for T21 and T18 was defined as >1/300 and >1/350, respectively.
Karyotyping
Cytogenetic analyses were performed as described previously (2). After amniocentesis, two technicians independently performed karyotyping using the GSL-120 instrument (Leica Biosystems Richmond, Inc.) and CytoVision Automated Cytogenetics Platform software. At least five cell karyotypes were analysed, and 20 karyotypes were counted. In cases of chromosome mosaicism, 60–100 karyotypes were evaluated.
Prenatal CMA testing
From January 2015, we performed prenatal CMA testing in pregnant women who consented. Amniotic fluid (10 mL) was collected, and genomic DNA was extracted using a QIAamp DNA Blood Mini Kit (Qiagen Inc., Valencia, CA), amplified (250 ng), labelled, and hybridised to a CytoScan HD array platform (Affymetrix) according to the manufacturer’s protocol. A single nucleotide polymorphism (SNP) array test was performed using a CytoScan 750K microarray chip (Affymetrix). After hybridisation, the chip was washed with buffer and scanned using a laser scanner. The data were analysed using Chromosome Analysis Suite ver. 3.0 (ChAs) software.
Detection of chromosomal abnormalities by NIPT
We analysed the sensitivity of NIPT according to the detection level of NIPT (2) and prior reports. The detection rates (DRs) for T21, T18, and T13 were obtained from our clinical data (2) and two meta-analyses (3,14), and we calculated the mean detection rate (Table 1). NIPT is capable of detecting 99.1%, 98.5%, and 96.5% of T21, T18, and T13 abnormalities, respectively. For foetal sex chromosomal aneuploidies (SCAs), we adopted the mean value of two reports (3,14). NIPT is capable of detecting 94.4% of 45,X SCAs and 100% of other SCAs. We obtained the DRs of structural rearrangement, triploidy, mosaicism, and submicroscopic chromosomal abnormalities from prior reports (15,16). The DR of SNP-based NIPT for 22q11.2 deletion syndrome was 97.8%.
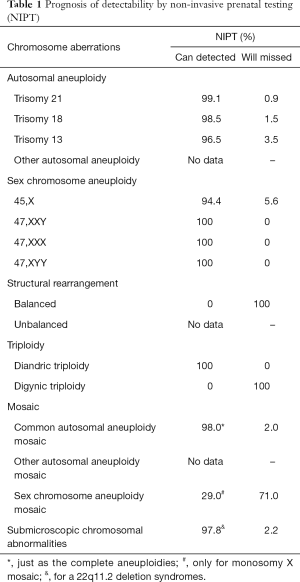
Full table
Results
In all, 3,099 women at high risk according to serological prenatal screening were analysed, comprising 2,857, 219, 15, and 8 at high risk for T21, T18, NTD, and both T21 and T18, respectively. After prenatal genetic counselling, the women underwent prenatal cytogenetic testing with amniocentesis (2,759 karyotyping, 59 karyotyping and CMA, and 281 CMA only).
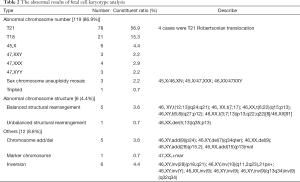
In all, 177 (5.7%) chromosomal abnormalities were found, comprising 129 (72.9%) abnormal chromosomes number, 6 (3.4%) abnormal chromosome structures, and 42 (23.7%) other abnormalities, including CNVs, inversion, and chromosome addition/deletion. From 2015, we used CMA for prenatal diagnosis. We detected a total of 27 (8.7%) CNVs in this study.
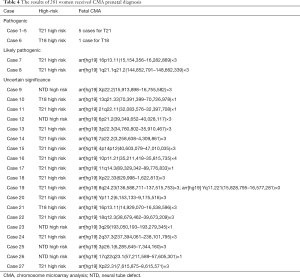
Among the 2,759 women who underwent only karyotyping, 137 (5.0%) abnormalities were detected, 86.9% (119/137) of which were an abnormal chromosome number (Table 2). Foetal autosomal aneuploidies (T21, T18, and T13) were the most prevalent type of abnormality, accounting for 72.3% (99/137) of the total. Among the 59 women who underwent both karyotyping and CMA, 50 showed consistent results (3 abnormalities) and 9 showed inconsistent results (5 abnormal CMA but normal karyotyping results and 4 normal CMA but abnormal karyotyping results) (Table 3). Among the 281 women who underwent only CMA testing, 6 had pathogenic CNVs, 2 had likely pathogenic CNVs, and 19 had CNVs of uncertain significance (Table 4).
Full table

Full table
Full table
Of the 177 (70.0%) abnormalities identified by cytogenetic testing, 124 could be detected by NIPT (Table 5). Of the 129 abnormal number of chromosomes, 124 and 5 (96.1% and 3.9%) were detected and missed by NIPT, respectively.
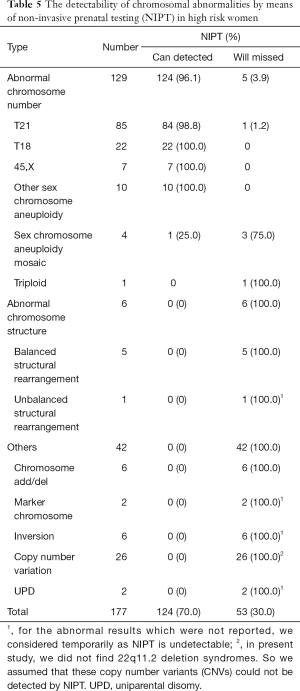
Full table
Discussion
Invasive prenatal diagnosis is used after serological screening in pregnant women at high risk for developing foetuses with chromosomal abnormalities. In recent years, NIPT has transformed prenatal screening and diagnosis as it has a high DR for T21, T18, T13, and some sex chromosome diseases. However, despite its accuracy, NIPT also has several limitations.
It has a sensitivity of >90% and a low false positive rate for detection of T21, T18, and T13. For common foetal aneuploidies and SCAs, it has a false positive rate of 0.09% (17) and 1% (1), respectively. The reasons for these rates are unclear but may be related to confined placental mosaicism, maternal mosaicism, vanished twins, maternal tumours, and/or maternal CNVs. However, only 33% of false-positive results for autosomal trisomy are reportedly related to biological or technical factors (18).
NIPT was developed to detect T21, T18, and T13, the focus of prenatal screening and diagnosis. However, advancements in technology have enabled the detection of other foetal diseases, which is necessary to prevent birth defects. Based on a 10-year experience of first-trimester screening, Alamillo et al. reported that almost 30% of foetal chromosomal abnormalities were beyond common trisomies (19), including trisomy 16, 47,XYY, 45,X, triploidy, and polyploidy. In this study, T21/T18/T13 accounted for 60.5% of the foetal chromosomal abnormalities. Therefore, if invasive prenatal diagnostic methods were replaced by NIPT, the frequency of invasive procedures would be reduced but 30.0% of abnormalities would be missed; this is in accordance with a prior study (20).
Most cases of an abnormal number of chromosomes were detected by NIPT. Similarly, Maxwell et al. (21) reported that NIPT detects about 85% of foetal karyotype abnormalities in women identified as high risk in first trimester screening. Srebniak et al. estimated that about 2–10% of foetuses with nuchal translucency would be missed by NIPT (22), as would a proportion of chromosome structural rearrangements, mosaicisms, triploides, and CNVs. This was likely due to amplification of foetal cfDNA from maternal plasma and MPS, resulting in mixing and partial degradation of foetal and maternal DNA. However, NIPT is not suitable for detecting chromosome structural abnormalities, for which amniocentesis or chorionic villus sampling is required. Similarly, NIPT is not useful for detecting CNV. Structural abnormalities (in particular, imbalanced rearrangements) of chromosomes do not alter their DNA content, and foetal mosaicism and CNV have a negligible impact on their DNA content, rendering NIPT largely useless. An enhanced NIPT method (NIPT-Plus) has shown good positive predictive values for microdeletion/microduplication syndromes (e.g., DiGeorge syndrome, 22q11.22 microduplication, Prader–Willi syndrome, Angelman syndrome, and Cri du Chat syndrome) (23). Cell-based NIPT is capable of detecting a variety of foetal chromosomal diseases, including aneuploidies, mosaicism, and unbalanced translocation (24). In summary, NIPT is capable of detecting T21/T18/T13 and several sex chromosome diseases, but not structural abnormalities, triploidy, mosaicism, microdeletions, or microduplications. Most of these non-detectable abnormalities are non-fatal but impact reproductive health in the long term by increasing the risk of infertility and abortion.
The DR of NIPT (70.0%) was lower than reported previously (25), probably due to the application of prenatal CMA. In this study, 11.0% (340/3,099) of the women underwent CMA, and this number increased after clinical counselling. Compared to karyotyping, CMA provides additional clinically important information. According to Wapner et al., it increases the DR of genetic abnormalities by 1.7% (26) and is capable of detecting aneuploidies and unbalanced rearrangements but not balanced translocations and triploides (26). In addition, CMA, but not NIPT, can identify variants of unknown clinical significance (VUS), which induce stress and anxiety in pregnant women. Use of NIPT to detect non-clinically significant VUS would alleviate such stress. Thus, prenatal CMA would result in detection of foetal abnormalities that would be missed by NIPT. To date, NIPT has been used for prenatal detection of DiGeorge syndrome, 22q11.2 deletion, Prader–Willi syndrome, Angelman syndrome, and balanced translocation (16,23,27). Further development of NIPT will increase its clinical utility.
In conclusion, we calculated the ability of NIPT to detect chromosomal abnormalities in pregnant women at high risk. NIPT will miss about 30.0% of the chromosomal abnormalities detected by invasive prenatal diagnostic methods. However, further development of NIPT will improve its efficacy.
Acknowledgments
We thank all of the project participants for their contributions. We also thank Textcheck (http://www.textcheck.com) for language editing.
Funding: This study was supported by grants from Project supported by National Natural Science Foundation of China (81773438), Key research and development plan project of Jiangsu Province (BE2017650, BE2018677), Changzhou science and technology support project (Social Development CE20175021), Lianyungang science and technology support project (Social Development SH1542).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study design and protocol were reviewed and approved by the Ethics Committee of the Changzhou Maternity and Child Health Care Hospital. Written informed content was obtained from the women prior to screening and diagnosis.
References
- Zhang B, Lu BY, Yu B, et al. Noninvasive prenatal screening for fetal common sex chromosome aneuploidies from maternal blood. J Int Med Res 2017;45:621-30. [Crossref] [PubMed]
- Yu B, Lu BY, Zhang B, et al. Overall evaluation of the clinical value of prenatal screening for fetal-free DNA in maternal blood. Medicine (Baltimore) 2017;96:e7114. [Crossref] [PubMed]
- Gil MM, Accurti V, Santacruz B, et al. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol 2017;50:302-14. [Crossref] [PubMed]
- Gil MM, Quezada MS, Revello R, et al. Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol 2015;45:249-66. [Crossref] [PubMed]
- Norton ME, Jacobsson B, Swamy GK, et al. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med 2015;372:1589-97. [Crossref] [PubMed]
- Olivieri S, Conti A, Iannaccone S, et al. Ceruloplasmin oxidation, a feature of Parkinson's disease CSF, inhibits ferroxidase activity and promotes cellular iron retention. J Neurosci 2011;31:18568-77. [Crossref] [PubMed]
- Advani HV, Barrett AN, Evans MI, et al. Challenges in non-invasive prenatal screening for sub-chromosomal copy number variations using cell-free DNA. Prenat Diagn 2017;37:1067-75. [Crossref] [PubMed]
- American College of Obstetricians and Gynecologists. Cell-free DNA screening for fetal aneuploidy. Committee Opinion No. 640. Obstet Gynecol 2015;126:e31-7. [Crossref] [PubMed]
- Benn P, Borell A, Chiu R, et al. Position statement from the Aneuploidy Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenat Diagn 2013;33:622-9. [Crossref] [PubMed]
- Gregg AR, Skotko BG, Benkendorf JL, et al. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genetics in Medicine 2016;18:1056-65. [Crossref] [PubMed]
- Choy KW, Kwok YK, Cheng YK, et al. Diagnostic accuracy of the BACs-on-Beads™ assay versus karyotyping for prenatal detection of chromosomal abnormalities: a retrospective consecutive case series. BJOG 2014;121:1245-52. [Crossref] [PubMed]
- Levy B, Wapner R. Prenatal diagnosis by chromosomal microarray analysis. Fertil Steril 2018;109:201-12. [Crossref] [PubMed]
- Ganapathi M, Nahum O, Levy B. Prenatal Diagnosis Using Chromosomal SNP Microarrays. Methods Mol Biol 2019;1885:187-205. [Crossref] [PubMed]
- Mackie FL, Hemming K, Allen S, et al. The accuracy of cell-free fetal DNA-based non-invasive prenatal testing in singleton pregnancies: a systematic review and bivariate meta-analysis. BJOG 2017;124:32-46. [Crossref] [PubMed]
- Shani H, Goldwaser T, Keating J, et al. Chromosomal abnormalities not currently detected by cell-free fetal DNA: a retrospective analysis at a single center. Am J Obstet Gynecol 2016;214:729.e1-11. [Crossref] [PubMed]
- Wapner RJ, Babiarz JE, Levy B, et al. Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. Am J Obstet Gynecol 2015;212:332.e1-9. [Crossref] [PubMed]
- Lutgendorf MA, Stoll KA, Knutzen DM, et al. Noninvasive prenatal testing: limitations and unanswered questions. Genet Med 2014;16:281-5. [Crossref] [PubMed]
- Hartwig TS, Ambye L, Sørensen S, et al. Discordant non-invasive prenatal testing (NIPT) - a systematic review. Prenat Diagn 2017;37:527-39. [Crossref] [PubMed]
- Alamillo CM, Krantz D, Evans M, et al. Nearly a third of abnormalities found after first-trimester screening are different than expected: 10-year experience from a single center. Prenat Diagn 2013;33:251-6. [Crossref] [PubMed]
- Lindquist A, Poulton A, Halliday J, et al. Prenatal diagnostic testing and atypical chromosome abnormalities following combined first-trimester screening: implications for contingent models of non-invasive prenatal testing. Ultrasound Obstet Gynecol 2018;51:487-92. [Crossref] [PubMed]
- Maxwell S, Dickinson JE, Murch A, et al. The potential impact of NIPT as a second-tier screen on the outcomes of high-risk pregnancies with rare chromosomal abnormalities. Aust N Z J Obstet Gynaecol 2015;55:420-6. [Crossref] [PubMed]
- Srebniak MI, de Wit MC, Diderich KE, et al. Enlarged NT (≥3.5 mm) in the first trimester - not all chromosome aberrations can be detected by NIPT. Mol Cytogenet 2016;9:69. [Crossref] [PubMed]
- Liang D, Cram DS, Tan H, et al. Clinical utility of noninvasive prenatal screening for expanded chromosome disease syndromes. Genet Med 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Vestergaard EM, Singh R, Schelde P, et al. On the road to replacing invasive testing with cell-based NIPT_ Five clinical cases with aneuploidies, microduplication, unbalanced structural rearrangement, or mosaicism. Prenat Diagn 2017;37:1120-4. [Crossref] [PubMed]
- Norton ME, Currier R, Jelliffe-Pawlowski L. Rare chromosome abnormalities detected by current prenatal screening compared to expected performance using non-invasive prenatal testing (NIPT). Am J Obest Gyneco 2014;210:S3-4. [Crossref]
- Wapner RJ, Martin CL, Levy B, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med 2012;367:2175-84. [Crossref] [PubMed]
- Jensen TJ, Kim SK, van den Boom D, et al. Noninvasive detection of a balanced fetal translocation from maternal plasma. Clin Chem 2014;60:1298-305. [Crossref] [PubMed]


