
Two novel mutations of PEX6 in one Chinese Zellweger spectrum disorder and their clinical characteristics
Introduction
Zellweger spectrum disorder (ZSD) is a subtype of peroxisome biogenesis disorder (PBD) caused by defects in peroxisomal biogenesis proteins. ZSD was used to be classified into Zellweger syndrome (ZS), neonatal adrenoleukodystrophy (NALD) and infantile Refsum disease (IRD), among which ZS was the most severe form and IRD was the least (1). Recently, this classification has been updated by regarding ZSD as an overall spectrum disease, with clinical features from severe, intermediate to mild phenotype (2). In addition, Heimler syndrome, another recessive PBD with mild clinical manifestations, is included in the ZSD spectrum as well. Clinical manifestations of ZSD range from dysmorphic features, sensory deficits to hypotonia, spastic ataxia, and life-threatening problems in multiple organs.
Bi-allelic mutations in any of the 13 PEX family genes can lead to ZSD, among which mutations in PEX1 and PEX6 are the most frequent, accounting for 60–70% and 10–15% respectively (3). Both PEX1 and PEX6 encode members of AAA (ATPases associated with diverse cellular activities) family of ATPases and form complexes anchored to the peroxisomal membrane by interaction with PEX26, a peroxisomal membrane protein. Pathogenic mutations in either PEX1 or PEX6 will cause export deficits of PEX5, a peroxisomal matrix protein receptor important for peroxisomal matrix protein import (4). This results in the damage of peroxisome-dependent metabolism pathway leading to accumulation or shortage of metabolites such as very long chain fatty acids (VLCFAs), C27-bile acid intermediates and erythrocyte plasmalogens (1).
Here we reported a Chinese female patient with PBD-ZSD due to two novel compound heterozygous mutations of p.Cys358* and p.Leu83Pro in PEX6. The phenotype-genotype association of PEX6 in PBD-ZSD is also reviewed.
Methods
Clinical material
Clinical materials of the proband were collected. Biochemistry analysis and cranial magnetic resonance imaging (MRI) were carried out. The diagnosis of PBD-ZSD was made by the combination of clinical evaluations and genetic testing.
Genetic test
Genomic DNA was isolated from peripheral blood sample of the patient through a standard method (Qiagen, German). Leukoencephalopathy gene panel sequencing was performed by target sequencing of the exons. In brief, all exons and their corresponding flanking regions of these genes were selected as target regions. Paired-end sequencing was performed on Illumina HiSeq X-ten platform. All variants different from the reference sequence were further screened by allele frequency <1% according to 1000 Genomes Project (http://www.internationalgenome.org/data), Inhouse database, ESP6500 (evs.gs.washington.edu/EVS/) and ExAC (exac.broadinstitute.org). The synonymous variants were excluded. The phenotypes of the screened genes were compared with the clinical manifestations of the proband and the inherited modes were considered to further exclude irrelevant genes. The mutations left were confirmed by Sanger sequencing. SIFT (5), Provean (6), Polyphen2 (7), Mutation Taster (8) and MUpro (9) were used to predict the pathogenicity of the variants.
Interpretation of the variants was based on the American College of Medical Genetics and Genomics (ACMG) recommended standards (10).
Updated review of the literature
An updated review was performed to identify primary articles reporting individuals of PBDs due to mutations in PEX6. As the diseases associated with PEX6 include both ZSD and Heimler syndrome (11), a rare kind of PBDs, a PubMed search with search terms of (“PEX6” OR “PAF-2”) AND ((“ZSD” OR “Zellweger Spectrum Disorder” OR “ZSS” OR “Zellweger Spectrum Syndrome”) OR (“PBD” OR “peroxisome biogenesis disorder”) OR (“Heimler Syndrome”) OR (“ZS” OR “Zellweger Syndrome”) OR (“NALD” OR “Neonatal Adrenoleukodystrophy”) OR (“IRD” OR “Infantile Refsum Disease”)) was performed. All studies included had to meet the following criteria: (I) containing patients who were diagnosed as PBDs with at least the mutation information and population were provided; (II) the pathogenicity is derived from PEX6 mutations rather than any other PEX genes. Secondary articles such as reviews were screened for additional information.
Results
Clinical evaluation
The patient was a 29-year-old female with flattened face and broad nasal bridge. She referred to Huashan Hospital after a five-year history of hand and head tremor as well as torticollis under no predisposing cause. The patient was born with retinitis pigmentosa and bilateral sensorineural hearing loss. Not only intellectual disability but ovarian and enamel dysplasia were shown during her growth and development. Family history was denied (Figure 1A). During her last physical examination, the patient displayed less cooperation, poor vision, weak light reflex of both eyes and a wide base gait. The muscle strength and muscle tone were normal. Biochemical analysis showed elevated VLCFAs level (C26:0 1.79 nmol/mL, normal range of ≤1.3 nmol/mL; C26:0/C22:0 =0.052, normal range of ≤0.023). Cranial MRI performed at age 24 indicated obvious brain atrophy and symmetrical subcortical and periventricular high signals (Figure 1B).
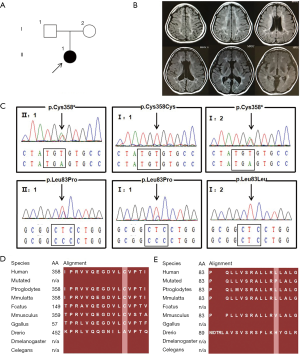
Mutation analysis
The mean depth of target sequencing of the patient was 84.780X. The percentage of the target regions with mean depth over 20X was 99.2%. Two heterozygous variants of c.1074T > A and c.248T > C in PEX6 (NM_000287.3) were found, leading to p.Cys358* and p.Leu83Pro change in exon3 and exon1 respectively (Figure 1C). No possible variants in another 12 PEX family genes were found. The variant was not detected in 200 Chinese seniors without medical history of neurological diseases from the community by Sanger sequencing. The parents of the patient were identified as heterozygous carriers of the two mutations separately by Sanger Sequencing (Figure 1C). Online bioinformatics prediction tools were used to estimate the pathogenicity of two variants (Table 1). Both variants have not been reported and were absent from ExAC database. In addition, according to the online protein analysis (http://www.cmbi.ru.nl/hope/), the mutant residue of p.Leu83Pro was considered to be smaller than the wide-type residue and disrupted a α-helix structure, which may consequently result in loss of interaction with other proteins. The wild-type residue was partially conserved at two positions (Figure 1D,E).

Full table
According to ACMG standards, both p.Cys358* (PVS1+PS3+PM2+PM4+PP3+PP4) and p.Leu83Pro (PS3 + PM2 + PM3 + PP3 + PP4) were classified as pathogenic variants (10). We diagnosed the patient as ZSD in combination of the clinical evaluations and genetic analysis.
Phenotype-genotype analysis of PBD patients with PEX6 mutation
Here, an updated review was performed to identify primary articles reporting individuals of PBDs due to mutations in PEX6. In all, we identified 16 articles containing 53 patients diagnosed as PBDs with PEX6 mutations (11-25) (http://fp.amegroups.cn/cms/atm.2019.06.42-1.pdf). Although ZSD was as an autosomal recessive inheritance disease, there were 11 ZSD patients showed the mode of autosomal dominant inheritance, which could be explained by allelic expression imbalance (13). In the remaining 42 patients, 47.6% showed homozygous mutations and 52.4% carried compound heterozygous mutations.
Among the fifty-three patients with ZSD, thirty-six were reported with clinical manifestations, of whom 61.1% displayed metabolism deficits including liver dysfunction and adrenal insufficiency. Other clinical characteristics include psychomotor development delay (50%), hypotonia (47.2%), vision disability (77.8%), sensorineural hear loss (66.7%), seizures (16.7%) and enamel dysplasia (36.1%).
In these patients who survived into adulthood (11,13,18,23), onset age was less than 3 years old, and the disease duration is usually over 18 years. Most of them have sensorineural hearing loss, pigmentary retinopathy and liver dysfunction. Other clinical manifestations include development delay, white matter abnormality, enamel dysplasia and facial dysmorphism. The patients with partial deficiency of peroxisomal functions may have normal reproductive capability (23). However, the patient in our case had displayed ovarian dysplasia. Since hypothalamic and pituitary lesions, Turner’s syndrome as well as toxic effect have been excluded according to the patient’s past medical history, we suspected ovarian dysplasia might be related to the PEX6 mutations, which needs to be further confirmed by more cases.
Discussion
We described a case of ZSD. The diagnosis was based on clinical characteristics of hypotonia, developmental delay, leukodystrophy and disease course, together with biochemical analysis and the detection of compound-heterozygous mutations of p.Cys358* and p.Leu83Pro in PEX6.
Disease caused by mutations of PEX6 include ZS, NALD, IRD and Heimler syndrome, all of which are now considered as the clinical continuums of PBD-ZSD, with the severity ranging from fatality to mildness (11). In our case, the patients displayed hand and head tremor as well as torticollis, which is consistence with the previous report by Kumar et al. in a patient with PEX16 mutations (26). Autosomal recessive cerebellar ataxia has been reported in patients with PEX16, PEX2 and PEX10 mutations (27-29) which was also noted in our patient. Besides, other neurological phenotypes such as peripheral neuropathy and progressive spastic paraparesis were not observed in our patient.
VLCFA is biosynthesized in the endoplasmic reticulum. The fatty chain of VLCFA is so long that it can only be metabolized in peroxisomes rather than mitochondria. Defects in multiple peroxisome enzyme pathways caused by mutations in PEX family lead to the accumulation of downstream fatty chains. According to our up-dated review, metabolism deficits including liver dysfunction and adrenal insufficiency is one of the most common phenotypes in PBD-ZSD patients with PEX6 mutations. Thus, screening of plasma metabolites such as VLCFAs can be a strong evidence for PBD-ZSD.
There are some limitations in this study. First, the functional assays to identify the pathogenicity of PEX6 mutations failed to be carried out. Second, since there is some missing information in the previous literature included for up-dated review, the genotype-phenotype correlation is not precise enough. A more comprehensive cohort study of PBD-ZSD is needed to further clarify the associations between genetics and clinical features.
Conclusions
In this article, we reported a Chinese PBD-ZSD patient with 2 novel compound heterozygous mutations of PEX6. Clinical evaluation and in silico analysis were performed. Since the variable combinations of the clinical manifestations in ZSD, Biochemical test such as VLFAC and Target sequencing was quite important in the diagnosis. We also found ovarian dysplasia in the patient but whether it is related to the PEX6 mutation should be further explored. As this is the first case of ZSD with PEX6 mutations in Chinese patient, further studies will focus on the contribution of PEX6 mutations to Chinese PBD patients and the pathogenicity of two novel mutations.
Acknowledgments
The authors sincerely appreciated the participants for their help and willingness to participate in this study.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was conducted after receiving written informed consent from the patient. This research was approved by the Institutional Ethics Committee of Huashan Hospital Fudan University. The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Braverman NE, D'Agostino MD, Maclean GE. Peroxisome biogenesis disorders: Biological, clinical and pathophysiological perspectives. Dev Disabil Res Rev 2013;17:187-96. [Crossref] [PubMed]
- Braverman NE, Raymond GV, Rizzo WB, et al. Peroxisome biogenesis disorders in the Zellweger spectrum: An overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol Genet Metab 2016;117:313-21. [Crossref] [PubMed]
- Ebberink MS, Mooijer PA, Gootjes J, et al. Genetic classification and mutational spectrum of more than 600 patients with a Zellweger syndrome spectrum disorder. Hum Mutat 2011;32:59-69. [Crossref] [PubMed]
- Grou CP, Carvalho AF, Pinto MP, et al. The peroxisomal protein import machinery--a case report of transient ubiquitination with a new flavor. Cell Mol Life Sci 2009;66:254-62. [Crossref] [PubMed]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4:1073-81. [Crossref] [PubMed]
- Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015;31:2745-7. [Crossref] [PubMed]
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248-9. [Crossref] [PubMed]
- Schwarz JM, Cooper DN, Schuelke M, et al. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014;11:361-2. [Crossref] [PubMed]
- Cheng J, Randall AZ, Sweredoski MJ, et al. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res 2005;33:W72-76. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Ratbi I, Falkenberg KD, Sommen M, et al. Heimler Syndrome Is Caused by Hypomorphic Mutations in the Peroxisome-Biogenesis Genes PEX1 and PEX6. Am J Hum Genet 2015;97:535-45. [Crossref] [PubMed]
- Wangtiraumnuay N, Alnabi WA, Tsukikawa M, et al. Ophthalmic manifestations of Heimler syndrome due to PEX6 mutations. Ophthalmic Genet 2018;39:384-90. [Crossref] [PubMed]
- Falkenberg KD, Braverman NE, Moser AB, et al. Allelic Expression Imbalance Promoting a Mutant PEX6 Allele Causes Zellweger Spectrum Disorder. Am J Hum Genet 2017;101:965-76. [Crossref] [PubMed]
- Rydzanicz M, Stradomska TJ, Jurkiewicz E, et al. Mild Zellweger syndrome due to a novel PEX6 mutation: correlation between clinical phenotype and in silico prediction of variant pathogenicity. J Appl Genet 2017;58:475-80. [Crossref] [PubMed]
- Smith CE, Poulter JA, Levin AV, et al. Spectrum of PEX1 and PEX6 variants in Heimler syndrome. Eur J Hum Genet 2016;24:1565-71. [Crossref] [PubMed]
- Matsunami M, Shimozawa N, Fukuda A, et al. Living-Donor Liver Transplantation From a Heterozygous Parent for Infantile Refsum Disease. Pediatrics 2016. [Crossref] [PubMed]
- Lusebrink N, Porto L, Waterham HR, et al. Absence of biochemical evidence at an early age delays diagnosis in a patient with a clinically severe peroxisomal biogenesis disorder. Eur J Paediatr Neurol 2016;20:331-5. [Crossref] [PubMed]
- Zaki MS, Heller R, Thoenes M, et al. PEX6 is Expressed in Photoreceptor Cilia and Mutated in Deafblindness with Enamel Dysplasia and Microcephaly. Hum Mutat 2016;37:170-4. [Crossref] [PubMed]
- Tran C, Hewson S, Steinberg SJ, et al. Late-onset Zellweger spectrum disorder caused by PEX6 mutations mimicking X-linked adrenoleukodystrophy. Pediatr Neurol 2014;51:262-5. [Crossref] [PubMed]
- Levesque S, Morin C, Guay SP, et al. A founder mutation in the PEX6 gene is responsible for increased incidence of Zellweger syndrome in a French Canadian population. BMC Med Genet 2012;13:72. [Crossref] [PubMed]
- Yik WY, Steinberg SJ, Moser AB, et al. Identification of novel mutations and sequence variation in the Zellweger syndrome spectrum of peroxisome biogenesis disorders. Hum Mutat 2009;30:E467-80. [Crossref] [PubMed]
- Krause C, Rosewich H, Thanos M, et al. Identification of novel mutations in PEX2, PEX6, PEX10, PEX12, and PEX13 in Zellweger spectrum patients. Hum Mutat 2006;27:1157. [Crossref] [PubMed]
- Raas-Rothschild A, Wanders RJ, Mooijer PA, et al. A PEX6-defective peroxisomal biogenesis disorder with severe phenotype in an infant, versus mild phenotype resembling Usher syndrome in the affected parents. Am J Hum Genet 2002;70:1062-8. [Crossref] [PubMed]
- Imamura A, Shimozawa N, Suzuki Y, et al. Temperature-sensitive mutation of PEX6 in peroxisome biogenesis disorders in complementation group C (CG-C): comparative study of PEX6 and PEX1. Pediatr Res 2000;48:541-5. [Crossref] [PubMed]
- Zhang Z, Suzuki Y, Shimozawa N, et al. Genomic structure and identification of 11 novel mutations of the PEX6 (peroxisome assembly factor-2) gene in patients with peroxisome biogenesis disorders. Hum Mutat 1999;13:487-96. [Crossref] [PubMed]
- Kumar KR, Wali G, Davis RL, et al. Expanding the spectrum of PEX16 mutations and novel insights into disease mechanisms. Mol Genet Metab Rep 2018;16:46-51. [Crossref] [PubMed]
- Sevin C, Ferdinandusse S, Waterham HR, et al. Autosomal recessive cerebellar ataxia caused by mutations in the PEX2 gene. Orphanet J Rare Dis 2011;6:8. [Crossref] [PubMed]
- Regal L, Ebberink MS, Goemans N, et al. Mutations in PEX10 are a cause of autosomal recessive ataxia. Ann Neurol 2010;68:259-63. [PubMed]
- Ebberink MS, Csanyi B, Chong WK, et al. Identification of an unusual variant peroxisome biogenesis disorder caused by mutations in the PEX16 gene. J Med Genet 2010;47:608-15. [Crossref] [PubMed]


