Phylogenetic analysis of clinical strains of Helicobacter pylori isolated from patients with gastric diseases in Tibet
Introduction
Helicobacter pylori (H. pylori) is a Gram-negative microaerophilic gastric pathogen, which could colonize in the upper gastrointestinal tracts over 50% population of the world (1). In 1982, H. pylori were isolated from gastric mucosa of a patient with chronic gastritis in Australia for the first time (2,3). Over the past three decades, a large number of studies on H. pylori have been carried out. It has shown that the infection of H. pylori is one of the major pathogenic reasons of chronic gastritis, peptic ulcer, gastric mucosa-associated lymphoid tissue (MALT) lymphoma and gastric cancer (4-6). The International Agency for Research on Cancer (IARC) of the World Health Organization (WHO) has defined H. pylori as class I carcinogen. Also, H. pylori can take up residence in the oral cavity and has been widely isolated from oral mucosa, plaque, and saliva of the oral disease’s sufferers (7,8). Oral H. pylori strains are associated with dental caries, periodontitis, mouth ulcers, ozostomia, and other oral diseases. H. pylori that were living in the oral cavity can be swallowed with saliva or food into the stomach, and the pathogen also can be vomited into the mouth from the stomach. However, the study on the oral H. pylori is scarce and far less than isolates from the stomach. The differences between the H. pylori strains that were isolated from the stomach and oral cavity are still controversial.
The initial infection of H. pylori usually occurs in childhood and always presents family aggregation (1,9,10). Without antibiotic eradication therapy, the infection of H. pylori in adults tends to be persistent lifelong. Many studies have shown that the prevalence of the infection of H. pylori was positively correlated with age (11,12). It is reported that the incidence and prevalence of the infection of H. pylori are common in China, especially in Tibet (11,13). An epidemiological investigation in 2010 (14) showed that the total infection rate of H. pylori in Chinese natural population was 56.22%, and it also showed significant differences among different areas of China. The lowest infection rate was 42.01% in Guangdong Province, while the highest was 84.62% in the Tibet Autonomous Region. It is noteworthy that Tibet is a high incidence of gastric cancer mortality area (13). According to the first gastric cancer census of Tibet in the 1970s, it suggested that the highest mortality of malignant tumors was gastric cancer, which accounted for 48.89% of total fatality of malignant tumors. Since then, gastric cancer mortality in Tibet has remained high always caused most cancer deaths despite a slight decline (15,16). Although backward economic development and poor hygiene conditions are confounding factors for H. pylori infection, the H. pylori strain epidemic in Tibet may present high pathogenicity and carcinogenicity.
Multi-locus sequence typing (MLST) is a rapid and effective method that based on allelic variation among several conserved housekeeping genes and has been applied to categorize and trace epidemic isolates of many pathogenic bacteria species for nearly two decades. In 2003, an MLST protocol for H. pylori employing seven housekeeping genes (ureI, mutY, EFP, PPA, yphC, atpA, and trpC) and one virulence-associated gene (vacA) was established, and this protocol divided 370 H. pylori strains all over the world into seven populations and subpopulations on the basis of their geographical distribution (17). In recent years, next generation sequencing (NGS) has been widely used in studying the genomics of bacteria and can provide much more detailed and precise genetic information (18). Until now, a large number of MLST typing and genome sequences of H. pylori strains have been obtained and published on a public database. However, there is only one H. pylori strain that isolated from a Tibetan patient with gastric cancer and was conducted genome sequencing. The similarity to the reference strain H. pylori 26695 was only 87.25% (16).
Tibet Autonomous Region is located in the Qinghai-Tibet Plateau, where the average elevation is over 4,000 meters. Unique geographical environment and weather conditions created a series of unique species distributions. The strain of H. pylori epidemic in Tibet may be more pathogenic and carcinogenic to maintain the high incidence of H. pylori infection and gastric cancer.
In this work, we intended to study the characteristics and structure of the epidemic strains of H. pylori in Tibet. To achieve this purpose, five H. pylori clinical strains in Tibet were isolated, identified and performed the genome sequencing. By comparing the genomic sequences of H. pylori 26695, the single nucleotide variants (SNVs) and Insertion-Deletions (InDels) were analyzed to learn more specificity of Tibetan strains. Also, 76 worldwide strains were performed phylogenetic analysis to study bacterial relationship and explore the population structure of the five epidemic strains of H. pylori in Tibet.
Methods
Isolation, culture, and identification of H. pylori strains
The study participants were recruited in gastroscopy room of Hospital of Chengdu Office, People’s Government of Tibet Autonomous Region during the period from December 2015 to February 2016. All participants did not have systemic diseases, and no antibiotics were used in one week before gastroscopy. Gastric mucosal tissues and saliva samples were collected for isolation of H. pylori clinical strains. Briefly, the gastric mucosal and salivary samples were tested by Urease Test. The positive samples were subsequently cultured on Columbia agar plates (Land Bridge, Beijing, China) with 5% sheep blood, at 37 °C in a microaerophilic environment containing 5% O2, 10% CO2, and 85% N2 for 72 h. After colonies appeared, gram stain and microscopic examination were carried out for initial identification; the bacterial colony was confirmed by a series of biochemical tests, such as Catalase, Oxidase and Urease Tests for further identification. H. pylori animal type strain, Sydney Strain 1 (SS1), was also cultured on Columbia agar base with 5% sheep blood as described above.
Five clinical strains were isolated from three patients with gastric diseases (Table 1). Of these, H. pylori XZCB-W009, XZCB-K00901 and XZCB-K00902 were isolated from patient No. 9, a 30-year-old male Tibetan patient with gastric ulcer, as XZCB-W009 was isolated from the stomach, XZCB-K00901 and XZCB-K00902 were from the oral cavity. XZCB-K002 was isolated from the stomach of patient No. 2, a 74-year-old female Han patient living in Tibet with gastritis. XZCB-K038 was isolated from the stomach of patient No. 38, a 75-year-old female patient with gastritis.

Full table
DNA extraction and Illumina sequencing
H. pylori strains were harvested from Columbia agar plates for DNA extraction, which was carried out by using TIANamp Bacteria DNA Kit (TIANGEN, Beijing, China) according to manufacturer’s instructions. The genomic DNA was then sequenced on a sequencer of Illumina Hiseq (2×150 bp) at GENEWIZ Inc. (Suzhou, China). The low quality reads were filtered by using Trimmomatic (v 0.30) (19) and Cutadapt (v 1.9.1) (20). The clean data were aligned to the reference genome of sequenced strain H. pylori 26695 (Accession Number NC_CP010436.1) by using BWA (v 0.7.12) (21). Identification of SNVs/InDels for each base site was performed by using SAMSTOOL (v 1.1) (22). The Annovar software was used for functional annotation on gene loci variation (23).
The genome sequences of these five Tibetan strains reported in this paper have been uploaded to the NCBI Sequence Read Archive (SRA) database under the BioProject SRP127752 with BioSample accession number SRS2800546, SRS2800545, SRS2800544, SRS2800548, and SRS2800547. The datasets generated during and analyzed during the current study are available in the SRA repository on the website of www.ncbi.nlm.nih.gov/sra/?term=SRP127752.
MLST analysis
For the MLST analysis, seven housekeeping genes (ureI, mutY, EFP, PPA, yphC, atpA, and trpC) and one virulence-associated gene (vacA) of five H. pylori strain from Tibet were selected and recombined as previously described (17,24). To analyze the phylogeny of the Tibet strains, eight gene fragments of the 76 H. pylori strains that isolated from 28 different countries across the five continents were downloaded from MLST database http://www.mlst.net (Table 2). Next, the ClustalX (v 1.81) was used for multiple sequence alignment of MLST genes (25). The alignments were used for phylogenic analysis with MEGA (v6.06) (26). Distance-based phylogenetic tree for MLST genes was calculated by using the Tamura-Nei model, and Bootstrap analysis was performed with 1,000 replications.
Full table
cagA gene
The cagA genes of five Tibetan strains were extracted from the genome sequence for phylogenetic analysis with strain 26695 and then were translated into protein sequence for EPIYA motifs detection.
Negative stain and electron microscope
H. pylori SS1, XZCB-W009, and XZCB-K00901 were cultured on Columbia agar plates for 72 h. The bacteria were suspended in 2% ammonium acetate to remove the medium, and stained by 2% sodium phosphotungstate. The samples were applied to copper Formvar-coated grids and observed under JEM-ARM 200F (JEOL) transmission electron microscope (TEM).
Results
Characteristics of H. pylori clinical strains
Gram stain and a series of biochemical tests were used for identification of H. pylori clinical strains. The H. pylori strains showed seagull or spiral shape under the microscope (data not shown). In biochemical tests, all the strains expressed positively for catalase, oxidase and urease tests (data not shown).
SNVs/InDels
In order to identify the genic mutations of the H. pylori strains from Tibet, genomes of five strains were sequenced via Illumina Hiseq platform. The sequencing harvested a 700× average depth and a >92% breadth coverage of the reference genome.
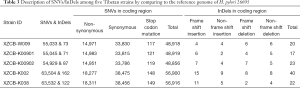
Genotypes of loci were ascertained by using the Bayesian algorithm to calculate the posterior probability of loci. Based on the differences between the calculated genetic loci and the reference genome of H. pylori 26695, each different genetic locus was predicated as SNVs or InDels. Among the five H. pylori clinical strains, 55,016–63,666 SNVs/InDels were detected and identified. Of which, there were approximately 89% of the SNVs/InDels located in the coding regions, and the remains were located in the non-coding regions, the downstream region and the intersection area of one upstream region and the other downstream region. It could be found that the genome sequences of all H. pylori strains were significantly different from the reference strain. The details of SNVs/InDels were shown in Table 3.
Full table
The SNVs located in the coding region were calculated and analyzed. Based on the functional alterations of the triplet codes, the SNVs were divided into three groups: synonymous mutation, non-synonymous mutation and stop codon mutation (referring to a stop codon mutates into a codon that encodes an amino acid, or an amino acid-encoding codon mutates into a stop codon). It was found that there were 33,786–38,475 and 14,951–18,311 SNVs in the synonymous group and non-synonymous group, accounted for 67.6–69.5% and 30.3–32.2% of the total SNVs numbers, respectively. Also, 116–149 SNVs, less than 0.3% of the total, were in stop codon mutation group (Table 3).
Based on breadth alteration of the open reading frames (ORFs) attributed to the insertion or deletion of the sequence fragments, the InDels in coding region were divided into four groups: non-frame shift insertion, frameshift insertion, non-frame shift deletion, and frameshift deletion. There were 17–40 InDels in the H. pylori clinical and type strains, comprising 2–9 non-frame shift insertions, 4–15 frameshift insertions, 4–13 non-frame shift deletions and 2–8 frameshift deletions (Table 3).
MLST analysis
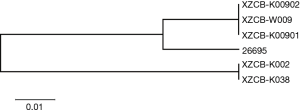
Fragments of seven house-keeping genes and one virulence-associated gene of each H. pylori strain were extracted and recombined. Among the 3,850 nucleotides, 1,250 were polymorphic and were used for phylogenetic analyses. Maximum likelihood unrooted tree diagram by MLST (Figure 1) showed that a total of 81 isolates were assigned into four groups: Asia, Oceania, Europe, and Africa group. Based on the molecular evolutionary relationship, all these isolates are defined by geography position and existing population.

H. pylori strain with defined Subpopulations hspEAsia, hpAsia 2, hspMario and hspAmerind were classified into Asia group. There were 18 strains that isolated from Beijing, Chongqing, Guangzhou, Hangzhou, Heilongjiang, Hubei, Shijiazhuang, Xi’an and Zhejiang of China, and two strains from Japan without a definition of structure population or subpopulation, which were assigned into Asia group. However, there were two strains isolated from Malaysia and Cambodia, which were identified as hpEurope. Another hpEurope strain from Sweden was located on a small branch next to the centric node in the Asia group.
A majority of isolates from Australia and New Zealand were assigned into Oceania group, an hpEurope population cluster. Interestingly, five Tibetan isolates were also classified in this cluster; H. pylori XZCB-K002 from a Han in Tibet and XZCB-K038 from a Tibetan were clustered in one branch with a Russian strain y1, regardless of race. XZCB-W009, XZCB-K00901, and XZCB-K00902 that from the same patient were clustered in one branch next to an Australian isolate NCTC 11637. Nearly all strains from Continental Europe with the definition of hpEurope were assigned into Europe group. All African strains were assigned in Africa group, which located in the farthest end from the centric node, but with one Italian strain be194.
cagA gene
The maximum likelihood tree for the cagA gene was shown in Figure 2. XZCB-W009, XZCB-K00901, and XZCB-K00902 isolated from the same patient were assigned in one cluster with strain 26695, XZCB-K002 and XZCB-K038 were clustered into another.
The EPIYA motifs in the C-terminal region of CagA protein, encoded by the cagA gene, were classified as Western type CagA-ABC (data not shown).
Electron microscopy of H. pylori
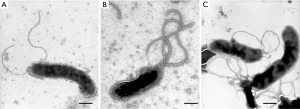
H. pylori SS1, XZCB-W009 and XZCB-K00901, were stained by negative staining and observed under TEM. The bacteria showed 2–6 flagella at one terminal of the body. The flagella of two clinical isolates were richer than SS1, and no significant differences were observed among XZCB-W009 and XZCB-K00901 (Figure 3).
Discussion
In this study, we analyzed the genome sequences of five H. pylori clinical strains isolated from three patients with gastric diseases in Tibet. Gene loci of SNVs and InDels were detected and identified in the genome of Tibetan strains by comparing to reference genome of H. pylori 26695. Based on the viewpoint of molecular genetic evolution, it is interesting finding that in our analysis with MLST, all five Tibetan strains presented more intimate with strains isolated from Australia and New Zealand and were quite different from other prevailing strains in other parts of China. Although Tibetan strains were assigned into the Oceania group and showed hpEurope ancestry, other Chinese strains were grouped as hspEAsia. This is the first study to explore the population structure of H. pylori strains epidemic in Tibet of China.
In previous reports, it is shown that according to the MLST analyze, Subpopulations of hspEAsia, hpAsia 2, hspMario and hspAmerind were assigned in the Asia group (27,28). Similarly, subpopulations of hpNEAfrica, hspWAfrica, and hspSAfrica were in the Africa group. In our study, the hpEurope population was split into the Oceania and Europe group. It is noteworthy that five H. pylori strains from Tibet were all in the Oceania group and defined as hpEurope. However, a previous Tibetan strain XZ274 isolated from a patient with gastric cancer presented a close relationship with strains isolated from southwest China (16,29).
The five Tibetan strains were cagA-positive strains. CagA, the product of the cagA gene, is delivered into gastric epithelial cells by Type IV Secretion System (TFSS). After tyrosine phosphorylation occurs, CagA specifically binds to SHP-2 phosphatase to activate to phosphatase activity and induce morphological transformation of cells (30). Polymorphism of EPIYA motifs in the C-terminal region of the cagA gene leads to pathogenic differences. East Asian CagA is characterized by the tandem arrangement of EPIYA-A, EPIYA-B and EPIYA-D segments in C-terminal region of CagA, whereas Western CagA contains EPIYA-A, EPIYA-B and an indefinite number (one to four) of EPIYA-C segments in tandem (31). The number of EPIYA-C segment is positively correlated with its pathogenicity (32). Accordingly, five strains from Tibet in this study were all defined as Western type CagA-ABC. Notably, East Asian CagA binds to SHP-2 more efficiently than Western type and synthesizes more virulence proteins (33). Moreover, strains of CagA-ABDD show more carcinogenicity than strains of CagA-ABCCC (34).
After isolating H. pylori strains from dental plaque of a dental caries patient for the first time in 1989 (35), the evolutionary relationship of H. pylori strains in the oral cavity and stomach has drawn much attention, but there is still no definite conclusion. Oral cavity, as an important colonization site of H. pylori, plays a significant role in the infection of H. pylori. There are differences in morphology and biochemical characteristics among H. pylori strains that isolated from different patients, whereas isolates from oral cavity and stomach of the same patient maintain a high-degree consistency (36). Subsequently, the homology of oral and gastric strains is also confirmed by molecular biological methods (37,38). In our phylogenetic analysis, it is confirmed that the gene sequence of three strains that were isolated from the oral cavity and the stomach of the same patient, XZCB-W009, XZCB-K00901, and XZCB-K00902, were almost identical. However, another study showed that from the same patient the agreement between oral and gastric genotypes was only 38.7% (39). In other studies, it is also shown that oral and gastric strains may be completely unrelated (40,41).
In conclusion, five H. pylori clinical strains from Tibet were isolated and performed genome sequencing. A huge amount of SNVs/InDels were detected and identified by comparing to H. pylori 26695. This study showed that the analyses of MLST improved the definition of H. pylori population particularly in Tibet. These Tibet strains were quite different from the strains that were isolated from other parts of China but showed hpEurope ancestry. Moreover, CagA of five Tibet strains showed Western type CagA-ABC, confirming this consequence. However, western CagA, due to less carcinogenic, did not seem to explain the phenomenon of the high prevalence of gastric cancer in the Tibet autonomous region. More Tibetan strains and researches are still needed for further investigation. Also, the relationship between oral and gastric strains from the same patient exhibited homology in molecular evolution.
Acknowledgments
Funding: This stud
y was supported by grants the Science Foundation of Tibet Autonomous Region (grant No. 2016ZR-15-2) and by Sichuan Province Science and Technology Project (grant No. 18ZDYF2323).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: All patients were required to provide written informed consent, and all procedures in the study were in complete accordance with the Helsinki Declaration and were reviewed and approved by the Ethics Committee of Hospital of Chengdu Office, People’s Government of Tibet Autonomous Region (Approval Number 2015-25).
References
- Khalifa MM, Sharaf RR, Aziz RK. Helicobacter pylori: a poor man's gut pathogen? Gut pathog 2010;2:2. [Crossref] [PubMed]
- Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1984;1:1311-5. [Crossref] [PubMed]
- Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1983;1:1273-5. [PubMed]
- Chmiela M, Karwowska Z, Gonciarz W, et al. Host pathogen interactions in Helicobacter pylori related gastric cancer. World J Gastroenterol 2017;23:1521-40. [Crossref] [PubMed]
- Fock KM, Graham DY, Malfertheiner P. Helicobacter pylori research: historical insights and future directions. Nat Rev Gastroenterol Hepatol 2013;10:495-500. [Crossref] [PubMed]
- Xu T, Fu D, Ren Y, et al. Genetic variations of TLR5 gene interacted with Helicobacter pylori infection among carcinogenesis of gastric cancer. Oncotarget 2017;8:31016-22. [PubMed]
- Aksit Bicak D, Akyuz S, Kiratli B, et al. The investigation of Helicobacter pylori in the dental biofilm and saliva samples of children with dyspeptic complaints. BMC oral health 2017;17:67. [Crossref] [PubMed]
- Castro-Munoz LJ, Gonzalez-Diaz CA, Munoz-Escobar A, et al. Prevalence of Helicobacter pylori from the oral cavity of Mexican asymptomatic children under 5 years of age through PCR. Arch Oral Biol 2017;73:55-9. [Crossref] [PubMed]
- Ignasi Elizalde J, Pique JM. Risk assessment in relatives of gastric cancer patients: hyperproliferation, genetics, and Helicobacter pylori infection. Eur J Gastroenterol Hepatol 2006;18:877-9. [Crossref] [PubMed]
- Nishizawa T, Suzuki H, Sakitani K, et al. Family history is an independent risk factor for the progression of gastric atrophy among patients with Helicobacter pylori infection. United European Gastroenterol J 2017;5:32-6. [Crossref] [PubMed]
- Asgeirsdottir GA, Kjartansdottir I, Olafsdottir AS, et al. Helicobacter pylori infection in Icelandic children. Scand J Gastroenterol 2017;52:686-90. [Crossref] [PubMed]
- Shi R, Xu S, Zhang H, et al. Prevalence and risk factors for Helicobacter pylori infection in Chinese populations. Helicobacter 2008;13:157-65. [Crossref] [PubMed]
- Zhao J, Geng P, Li Z, et al. Prostate stem cell antigen rs2294008 polymorphism differentially contributes to Helicobacter pylori-negative gastric cancer among various populations in China. Mol Clin Oncol 2013;1:493-8. [Crossref] [PubMed]
- Zhang W, Hu F, Xiao S, et al. Prevalence of Helicobacter pylori infection in China. Modern Digestion & Intervention 2010;15:265-70.
- Li K, Dan Z, Hu XJ, et al. Association of CD14/-260 polymorphism with gastric cancer risk in Highland Tibetans. World J Gastroenterol 2014;20:2688-94. [Crossref] [PubMed]
- Guo Y, Wang H, Li Y, et al. Genome of Helicobacter pylori strain XZ274, an isolate from a tibetan patient with gastric cancer in China. J Bacteriol 2012;194:4146-7. [Crossref] [PubMed]
- Falush D, Wirth T, Linz B, et al. Traces of human migrations in Helicobacter pylori populations. Science 2003;299:1582-5. [Crossref] [PubMed]
- Walker TM, Ip CL, Harrell RH, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 2013;13:137-46. [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet Journal 2011;17:10-2. [Crossref]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078-9. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [Crossref] [PubMed]
- Achtman M, Azuma T, Berg DE, et al. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol 1999;32:459-70. [Crossref] [PubMed]
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics 2002;Chapter 2:Unit 2.3.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013;30:2725-9. [Crossref] [PubMed]
- Dominguez-Bello MG, Perez ME, Bortolini MC, et al. Amerindian Helicobacter pylori strains go extinct, as European strains expand their host range. PloS One 2008;3:e3307. [Crossref] [PubMed]
- Munoz-Ramirez ZY, Mendez-Tenorio A, Kato I, et al. Whole Genome Sequence and Phylogenetic Analysis Show Helicobacter pylori Strains from Latin America Have Followed a Unique Evolution Pathway. Front Cell Infect Microbiol 2017;7:50. [Crossref] [PubMed]
- You Y, He L, Zhang M, et al. Comparative genomics of a Helicobacter pylori isolate from a Chinese Yunnan Naxi ethnic aborigine suggests high genetic divergence and phage insertion. PloS One 2015;10:e0120659. [Crossref] [PubMed]
- Higashi H, Tsutsumi R, Fujita A, et al. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci U S A 2002;99:14428-33. [Crossref] [PubMed]
- Hayashi T, Morohashi H, Hatakeyama M. Bacterial EPIYA effectors--where do they come from? What are they? Where are they going? Cell Microbiol 2013;15:377-85. [Crossref] [PubMed]
- Ferreira RM, Machado JC, Leite M, et al. The number of Helicobacter pylori CagA EPIYA C tyrosine phosphorylation motifs influences the pattern of gastritis and the development of gastric carcinoma. Histopathology 2012;60:992-8. [Crossref] [PubMed]
- Torres K, Valderrama E, Sayegh M, et al. Study of the oipA genetic diversity and EPIYA motif patterns in cagA-positive Helicobacter pylori strains from Venezuelan patients with chronic gastritis. Microb Pathog 2014;76:26-32. [Crossref] [PubMed]
- Miura M, Ohnishi N, Tanaka S, et al. Differential oncogenic potential of geographically distinct Helicobacter pylori CagA isoforms in mice. Int J Cancer 2009;125:2497-504. [Crossref] [PubMed]
- Krajden S, Fuksa M, Anderson J, et al. Examination of human stomach biopsies, saliva, and dental plaque for Campylobacter pylori. J Clin Microbiol 1989;27:1397-8. [PubMed]
- Young KA, Allaker RP, Hardie JM. Morphological analysis of Helicobacter pylori from gastric biopsies and dental plaque by scanning electron microscopy. Oral Microbiol Immunol 2001;16:178-81. [Crossref] [PubMed]
- Ferguson DA Jr, Li C, Patel NR, et al. Isolation of Helicobacter pylori from saliva. J Clin Microbiol 1993;31:2802-4. [PubMed]
- Oshowo A, Tunio M, Gillam D, et al. Oral colonization is unlikely to play an important role in Helicobacter pylori infection. Br J Surg 1998;85:850-2. [Crossref] [PubMed]
- Medina ML, Medina MG, Merino LA. Correlation between virulence markers of Helicobacter pylori in the oral cavity and gastric biopsies. Arq Gastroenterol 2017;54:217-21. [Crossref] [PubMed]
- Loster BW, Majewski SW, Czesnikiewicz-Guzik M, et al. The relationship between the presence of Helicobacter pylori in the oral cavity and gastric in the stomach. J Physiol Pharmacol 2006;57 Suppl 3:91-100. [PubMed]
- Lukes P, Pavlik E, Potuznikova B, et al. Comparison of Helicobacter pylori genotypes obtained from the oropharynx and stomach of the same individuals - a pilot study. Prague Med Rep 2012;113:231-9. [Crossref] [PubMed]