
SF3B1 mutation predicts unfavorable treatment-free survival in Chinese chronic lymphocytic leukemia patients
Introduction
Chronic lymphocytic leukemia (CLL) is a common kind of mature lymphocytic proliferative disease with heterogeneous clinical course. Although some patients can be “watch and wait,” a considerable number of patients need immediate treatment upon diagnosis and have diverse treatment responses and survival times. Multiple factors contribute to this heterogeneity (1-5). Age, β2-microglobulin (β2-MG) concentration, stage, tumor protein 53 (TP53) status, and immunoglobulin heavy variable-region gene (IGHV) status, compose the CLL-international prognostic index (CLL-IPI), which can stratify patients into 4 groups: low [0–1], intermediate [2–3], high [4–6], and very high risk [7–10]. Although CLL-IPI is widely used to assess the prognosis, novel prognosis-related genetic aberrations are not included in this system (6).
Splicing factor 3b subunit 1 (SF3B1), located in 2q33.1, encodes the subunit 1 of splicing factor 3b, a component of the U2 small nuclear ribonucleoproteins complex, which is crucial to RNA alternative splicing (7). SF3B1 mutation leads to the development and progression of multiple malignant diseases (8), including myelodysplastic syndromes (9), breast cancer (10), and CLL (11). In CLL, the most frequent aberration of SF3B1 is a missense mutation with a hotspot of c.A2098G (p.K700E) (4,12). The SF3B1 mutation occurs in 10–20% newly diagnosed CLL patients from western countries and suggests unfavorable prognosis (4,12). However, the incidence of SF3B1 mutation is relatively low in Chinese CLL patients, and its prognostic value remains controversial (5,13). In this study, we retrospectively analyzed the incidence and clinical impact of SF3B1 mutation in 399 newly diagnosed CLL patients. Additionally, new risk models based on SF3B1 mutational status and CLL-IPI were developed in order to predict the survival of Chinese CLL patients better.
Methods
Patients
A total of 399 previously untreated CLL patients [including 201 patients reported by Xia et al. (5)] diagnosed from January 2000 to December 2017 in our hospital were enrolled in this single-center retrospective study. Diagnosis of CLL was based on the International Workshop on CLL-National Cancer Institute criteria. The hospital ethics committee approved this study, and all patients were provided and signed informed consent according to the Declaration of Helsinki.
SF3B1 mutation detection
Genomic DNA from CLL samples was extracted as previously reported (5). Briefly, PCR amplification was performed for exon 14-16 of SF3B1. Primers used were as follows: SF3B1 exon 14 forward primer 5'-TGACTGTCCTTTCTTTGTTTAC-3' and reverse primer 5'-ATAGTAAGACCCTGTCTCCTA-3'; exon 15-16 forward primer 5'-TTGGCTGAATAGTTGATATATTGAGAG-3' and reverse primer 5'-AAACACTTTAAAATTCTGTTAGAACCA-3'. Sanger sequencing was performed in exon 14-16.
Data collection
Laboratory data such as absolute lymphocyte count (ALC), platelet count (PLT), hemoglobin (Hb), serum albumin (ALB) concentration, lactate dehydrogenase (LDH), thymidine kinase 1 (TK-1), and β2-MG were accessible from the hospital-based laboratory service within 24 h after the first admission.
Detection of TP53, NOTCH1, and MYD88 mutation, and IGHV mutation status were performed as previously described (14). The cut-off of 98% homology to germline was used to dichotomize IGHV mutational status.
Karyotype analysis of CLL cells was performed after CpG-oligodeoxynucleotide and interleukin-2 stimulation. Fluorescence in situ hybridization (FISH) was carried out to detect 17p deletion, 11q deletion, 13q deletion, and trisomy 12 according to the procedures described previously (5). CD38 and ZAP-70 were detected via flow cytometry, and the cut-off values for positivity were 30% and 20% respectively.
Statistical analyses
SPSS 23 (IBM Corporation, Armonk, NY, USA) and MedCalc Statistical Software version 15.2.2 (MedCalc Software bvba, Ostend, Belgium) were used to analyze data. Categorical variables were presented in percentage (%) and analyzed by the χ2 test. Overall survival (OS) was defined as the time from diagnosis to death or last follow-up, and treatment-free survival (TFS) was calculated as the time between diagnosis and first-line treatment. Survival curves were constructed by the Kaplan-Meier method, and the log-rank test was used for statistic associations. The Cox proportional hazards model was established to evaluate different factors at diagnosis on survival by univariate and multivariate analyses. For the multivariate analysis, we included variables whose P value is less than 0.05 during the univariate analysis. Receiver-operator characteristic (ROC) curve and corresponding area under the curve (AUC) were constructed to assess the predictive accuracy of CLL-IPI and new risk models, and the differences in AUCs were tested by a nonparametric approach developed by DeLong et al. (15). The Hosmer-Lemeshow goodness of fit test tested the calibration of risk models, and a calibration plot was drawn with observed events/total events and corresponding expected events/total events like the X axis and Y axis, respectively. P<0.05 was defined as a statistically significant value. Graphs were made by SPSS 23 and GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Patient characteristics
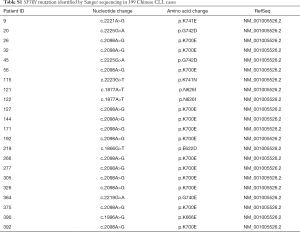
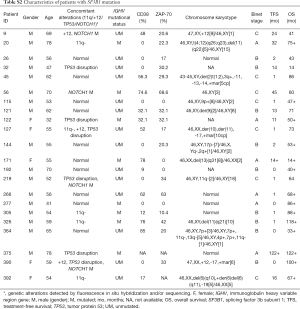
A total of 399 newly diagnosed Chinese CLL patients (261 male patients and 138 female patients) were enrolled in this study. Median age was 60 years old (16–93 years). There were 355 patients in Rai I-IV or Binet B/C. SF3B1 mutation was seen in 22 patients (5.5%) at sites of c.1866G>T, c.1877A>T, c.1996A>G, c.2098A>G, c.2219G>A, c.2221A>G, c.2223G>T, and c.2225G>A. The details of patients with SF3B1 mutation are shown in Tables S1,S2. The median follow-up time was 60 months (2–230 months). As of the last follow-up, 293 patients have been treated, and their regimens included chemoimmunotherapy (278/293) and ibrutinib (7/293). Information about treatment regimens are not available in 8 patients. Detailed information is presented in Table 1.
Full table
Full table

Full table
Clinical, cytogenetic and molecular associations
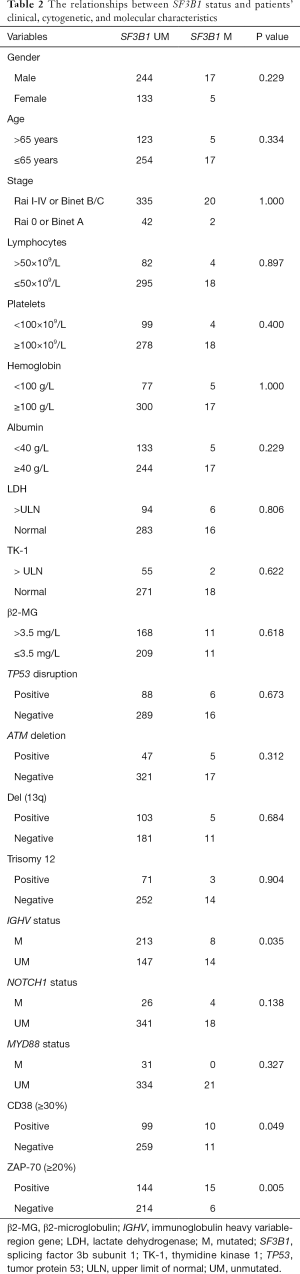
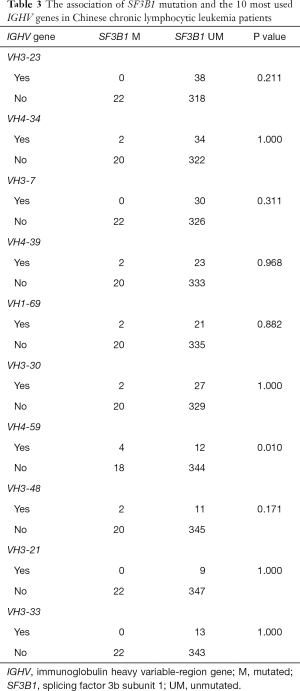
The relationships between SF3B1 mutation and patients’ clinical, cytogenetic, and molecular characteristics are shown in Table 2. SF3B1 mutation was common in patients with unmutated IGHV (P=0.035), positive CD38 (P=0.049), and positive ZAP-70 (P=0.005). Also, IGHV usage was available in 378 patients, and we analyzed the association of SF3B1 mutation with the 10 most used IGHV genes in Chinese CLL patients. A preferable IGHV 4-59 gene usage was observed in SF3B1-mutated subjects (Table 3).
Full table
Full table
Prognostic impact of SF3B1 mutation
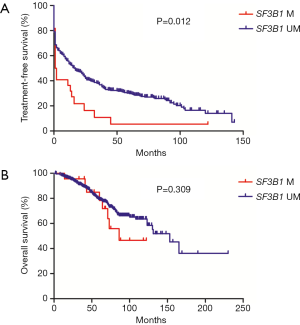
Patients with mutated SF3B1 had inferior TFS than those with unmutated SF3B1 (P=0.012), while no significant difference was seen in OS between the two groups (Figure 1). The 1- and 3-year TFS were 53.6% and 36.9% respectively, for SF3B1-unmutated patients vs. 36.4% and 10.9% respectively, for SF3B1-mutated patients.
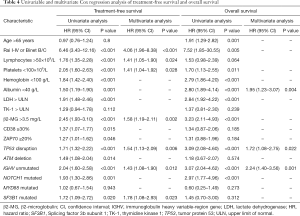
The results of Cox regression analysis are presented in Table 4. Univariate Cox regression analysis showed that Rai I-IV or Binet B/C (P<0.001), ALC >50×109/L (P<0.001), PLT <100×109/L (P<0.001), Hb <100 g/L (P<0.001), ALB <40 g/L (P=0.001), LDH > upper limit of normal (ULN) (P<0.001), β2-MG >3.5 mg/L (P<0.001), CD38 ≥30% (P=0.015), ZAP70 ≥20% (P=0.046), TP53 disruptions (P<0.001), ATM deletion (P=0.014), unmutated IGHV (P<0.001), mutated NOTCH1 (P=0.001) as well as mutated SF3B1 (P=0.020) had adverse impacts on TFS, and age >65 years (P=0.001), Rai I-IV or Binet B/C (P=0.005), PLT <100×109/L (P=0.011), Hb <100 g/L (P<0.001), ALB <40 g/L (P<0.001), LDH > ULN (P<0.001), β2-MG >3.5 mg/L (P<0.001), TP53 disruptions (P<0.001), unmutated IGHV (P<0.001), and mutated NOTCH1 (P<0.001) predicted shorter OS. Multivariate Cox regression analysis revealed that Rai I-IV or Binet B/C (P<0.001), ALC >50×109/L (P=0.024), PLT <100×109/L (P=0.028), β2-MG >3.5 mg/L (P=0.002), TP53 disruptions (P=0.006), unmutated IGHV (P=0.012), and mutated SF3B1 (P=0.023) were independently associated with inferior TFS, and ALB <40 g/L (P=0.004), TP53 disruptions (P=0.022), and unmutated IGHV (P=0.001) were independent prognostic factors for OS.
Full table
Subgroup analysis of SF3B1 mutation
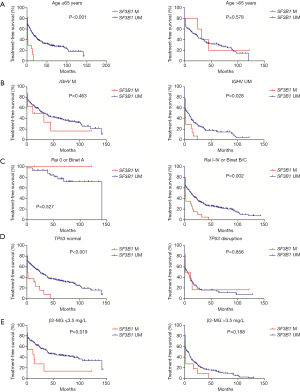
We divided patients into different subgroups according to age, IGHV mutation status, stage, TP53 status, and β2-MG concentration, and analyzed the prognostic impact of SF3B1 mutation in these subgroups (Figure 2). Worse TFS was observed in SF3B1-mutated patients in subgroups such as age ≤65 years old (P<0.001), IGHV unmutated (P=0.028), Rai I-IV or Binet B/C (P=0.002), TP53 normal (P<0.001), and β2-MG ≤3.5 mg/L (P=0.019).
New risk models based on SF3B1 mutation and CLL-IPI
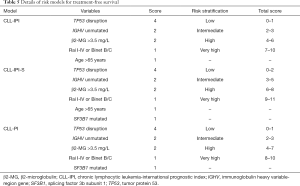
We developed two new risk models, CLL-IPI-S and CLL-PI, based on SF3B1 mutational status and CLL-IPI (Table 5). CLL-IPI-S consisted of 6 adverse variables, including TP53 disruptions, IGHV unmutated, β2-MG >3.5 mg/L, Rai I-IV or Binet B/C, age >65 years old, and SF3B1 mutated. Because age was not associated with adverse TFS in Chinese CLL patients, in the second system, we replaced age with SF3B1 mutation in CLL-IPI and named it CLL-PI. ROC curve was conducted to analyze the power of CLL-IPI, CLL-IPI-S, and CLL-PI in predicting TFS of Chinese CLL patients. For 6-month TFS (Figure 3A), the AUC of CLL-IPI, CLL-IPI-S, and CLL-PI were 0.664, 0.669, and 0.676 (CLL-IPI vs. CLL-PI, P=0.055), respectively. For 12-month TFS (Figure 3B), the AUC of CLL-PI was significantly larger than CLL-IPI (0.717 vs. 0.702, P=0.011) and CLL-IPI-S (0.717 vs. 0.705, P=0.027). For overall TFS, CLL-PI was a superior predictor for TFS than CLL-IPI (AUC: 0.773 vs. 0.758, P=0.029) and showed better tendency when compared with CLL-IPI-S (AUC: 0.773 vs. 0.762, P=0.096) (Figure 3C).
Full table
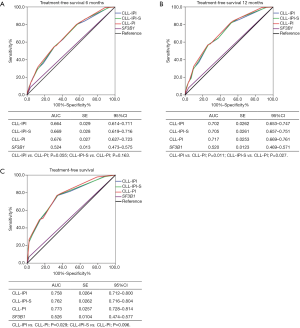
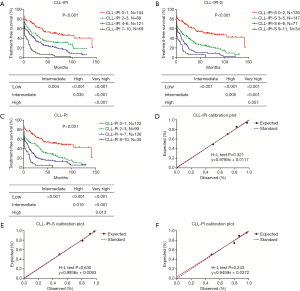
We further validated the potency of CLL-IPI-S (low-risk 0–2; intermediate risk 3–5; high-risk 6–8; very high-risk 9–11) and CLL-PI (low-risk 0–1; intermediate risk 2–3; high-risk 4–7; very high-risk 8–10) for predicting TFS in Chinese CLL patients (Table 5). Both CLL-IPI-S and CLL-PI could significantly separate Chinese CLL patients into 4 groups as CLL-IPI (P<0.001). Median TFS of low, intermediate, high, and very high-risk patients were 69, 25, 11, and 1 month for CLL-IPI, 69, 12, 6, and 1 month for CLL-IPI-S, and 79, 25, 8, and 1 month for CLL-PI (Figure 4). P value of the Hosmer-Lemeshow test was 0.321, 0.630, and 0.243 for CLL-IPI, CLL-IPI-S, and CLL-PI, respectively.
Discussion
Mutations affecting alternative splicing pathway genes play important roles in the pathogenesis and progression of CLL (7). SF3B1, encoding an important splicing factor, is frequently mutated in 10–20% of newly diagnosed CLL patients according to previous reports (12). SF3B1 mutation may lead to increased DNA damage, CLL-associated RNA alteration and Notch signaling activation through DVL2 alternative splicing, contributing to the poor prognosis of CLL patients harboring this mutation (11,16,17). In this study, we retrospectively analyzed the clinical, cytogenetic and molecular characteristics of 399 newly diagnosed Chinese CLL patients with different SF3B1 mutational status, explored the effects of SF3B1 mutation on their survival, and developed new risk models on the basis of CLL-IPI and SF3B1 mutational status to better predict the TFS of newly diagnosed CLL patients.
The incidence of SF3B1 mutation in Chinese CLL patients from our study was 5.5%, which was lower than that in patients from western countries. All mutations were missense, and the most common mutation was c.A2098G (p.K700E). Consistent with previous reports, SF3B1 mutation mainly occurred in patients with unmutated IGHV, positive CD38, and positive ZAP-70. Strefford et al. (18), Rossi et al. (19), and Jeromin et al. (20) reported that SF3B1 mutation showed a higher frequency in stereotyped IGHV3-21, IGHV3-48, and IGHV1-69 users. However, SF3B1 mutation in Chinese CLL patients was common in patients with IGHV4-59. Furthermore, in contrast to the results of western countries, SF3B1 mutation could only predict adverse TFS but failed to predict unfavorable OS in our cohort. Through subgroup analysis, we found that SF3B1 mutation could only represent adverse prognosis in subgroups such as age ≤65 years old, IGHV unmutated, Rai I-IV or Binet B/C, TP53 normal and β2-MG ≤3.5 mg/L.
These differences could be due to the following reasons. First, the differences of the clinical, cytogenetic and molecular background between CLL patients from Asian and western countries. Second, Nadeu et al. (12) reported that the incidence of SF3B1 mutation was 13% and nearly half the mutations were subclones with variant allele frequency <12%, which could not be detected by Sanger sequencing. Therefore, in our study, we missed some subclonal mutation due to Sanger sequencing, accounting for the lower prevalence of SF3B1 mutation in our cohort.
ROC curve was conducted to compare the power of TFS prediction of three risk models. Although the improvement was minor, CLL-PI had larger AUC than CLL-IPI and CLL-IPI-S with a significant statistical difference. Moreover, the calibration of CLL-PI was fairly satisfactory. However, SF3B1 mutation just represented adverse prognosis for TFS. Thus CLL-PI could only stratify Chinese patients in terms of TFS and had limited scope of application. Also, these risk models were developed based on a small Chinese cohort, and confirmation was needed in a larger cohort to estimate the repeatability of these models and judge the feasibility in patients from western countries.
So far, more than 13 studies have investigated the prognostic value of SF3B1 mutation in CLL (21). Most of them were from western countries, and only two studies investigated Asian populations. Our study focused on the effects of SF3B1 mutation on Chinese CLL patients, and the sample size was the largest among these Asian studies. Furthermore, our study was the first to combine SF3B1 mutation with CLL-IPI, and eventually improve the potency of CLL-IPI for predicting TFS in Chinese CLL patients.
In summary, we analyzed SF3B1 mutation in 399 newly diagnosed Chinese CLL patients. The incidence of SF3B1 mutation was 5.5% and was lower than that in previous studies from western countries. Also, SF3B1 mutation could only predict shorter TFS. New risk models were established according to the CLL-IPI and SF3B1 mutational status in order to better stratify Chinese patients in terms of TFS. Admittedly, there were some limitations in our study, such as small sample size, low-sensitivity sequencing, and short follow-up time. A larger study cohort and advanced next-generation sequencing technology are needed in order to identify the role of SF3B1 mutation further and confirm the potency of CLL-PI in Chinese CLL patients.
Acknowledgements
Funding: This study was supported by the National Natural Science Foundation of China (Grant No. 81370657, 81470328, 81600130, 81700193, 81770166, 81720108002), Jiangsu Province’s Medical Elite Program (ZDRCA2016022), the Natural Science Foundation of Jiangsu Province (BK20161354), the Excellent Youth Foundation Project of Jiangsu Province (BK20160099), the Project funded by Jiangsu Provincial Special Program of Medical Science (BE2017751), and the National Science and Technology Major Project (2018ZX09734007).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by the hospital ethics committee (2018-SRFA-087) of the First Affiliated Hospital of Nanjing Medical University, Jiangsu Province Hospital.
References
- Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011;475:101-5. [Crossref] [PubMed]
- Quijada-Alamo M, Hernandez-Sanchez M, Robledo C, et al. Next-generation sequencing and FISH studies reveal the appearance of gene mutations and chromosomal abnormalities in hematopoietic progenitors in chronic lymphocytic leukemia. J Hematol Oncol 2017;10:83. [Crossref] [PubMed]
- Quijano S, Lopez A, Rasillo A, et al. Impact of trisomy 12, del(13q), del(17p), and del(11q) on the immunophenotype, DNA ploidy status, and proliferative rate of leukemic B-cells in chronic lymphocytic leukemia. Cytometry B Clin Cytom 2008;74:139-49. [Crossref] [PubMed]
- Rasi S, Khiabanian H, Ciardullo C, et al. Clinical impact of small subclones harboring NOTCH1, SF3B1 or BIRC3 mutations in chronic lymphocytic leukemia. Haematologica 2016;101:e135-8. [Crossref] [PubMed]
- Xia Y, Fan L, Wang L, et al. Frequencies of SF3B1, NOTCH1, MYD88, BIRC3 and IGHV mutations and TP53 disruptions in Chinese with chronic lymphocytic leukemia: disparities with Europeans. Oncotarget 2015;6:5426-34. [Crossref] [PubMed]
- International CLL-IPI working group. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): a meta-analysis of individual patient data. Lancet Oncol 2016;17:779-90. [Crossref] [PubMed]
- Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer 2016;16:145-62. [Crossref] [PubMed]
- Obeng EA, Chappell RJ, Seiler M, et al. Physiologic Expression of Sf3b1(K700E) Causes Impaired Erythropoiesis, Aberrant Splicing, and Sensitivity to Therapeutic Spliceosome Modulation. Cancer Cell 2016;30:404-17. [Crossref] [PubMed]
- Malcovati L, Papaemmanuil E, Bowen DT, et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood 2011;118:6239-46. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70. [Crossref] [PubMed]
- Wang L, Brooks AN, Fan J, et al. Transcriptomic Characterization of SF3B1 Mutation Reveals Its Pleiotropic Effects in Chronic Lymphocytic Leukemia. Cancer Cell 2016;30:750-63. [Crossref] [PubMed]
- Nadeu F, Delgado J, Royo C, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood 2016;127:2122-30. [Crossref] [PubMed]
- Wu SJ, Lin CT, Agathangelidis A, et al. Distinct molecular genetics of chronic lymphocytic leukemia in Taiwan: clinical and pathogenetic implications. Haematologica 2017;102:1085-90. [Crossref] [PubMed]
- Wang WT, Zhu HY, Wu YJ, et al. Elevated absolute NK cell counts in peripheral blood predict good prognosis in chronic lymphocytic leukemia. J Cancer Res Clin Oncol 2018;144:449-57. [Crossref] [PubMed]
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837-45. [Crossref] [PubMed]
- Agrawal AA, Seiler M, Brinton LT, et al. Novel SF3B1 in-frame deletions result in aberrant RNA splicing in CLL patients. Blood Adv 2017;1:995-1000. [Crossref] [PubMed]
- Te Raa GD, Derks IA, Navrkalova V, et al. The impact of SF3B1 mutations in CLL on the DNA-damage response. Leukemia 2015;29:1133-42. [Crossref] [PubMed]
- Strefford JC, Sutton LA, Baliakas P, et al. Distinct patterns of novel gene mutations in poor-prognostic stereotyped subsets of chronic lymphocytic leukemia: the case of SF3B1 and subset #2. Leukemia 2013;27:2196-9. [Crossref] [PubMed]
- Rossi D, Bruscaggin A, Spina V, et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood 2011;118:6904-8. [Crossref] [PubMed]
- Jeromin S, Weissmann S, Haferlach C, et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia 2014;28:108-17. [Crossref] [PubMed]
- Zhang Z, Chen S, Chen S, et al. SF3B1 mutation is a prognostic factor in chronic lymphocytic leukemia: a meta-analysis. Oncotarget 2017;8:69916-23. [PubMed]


